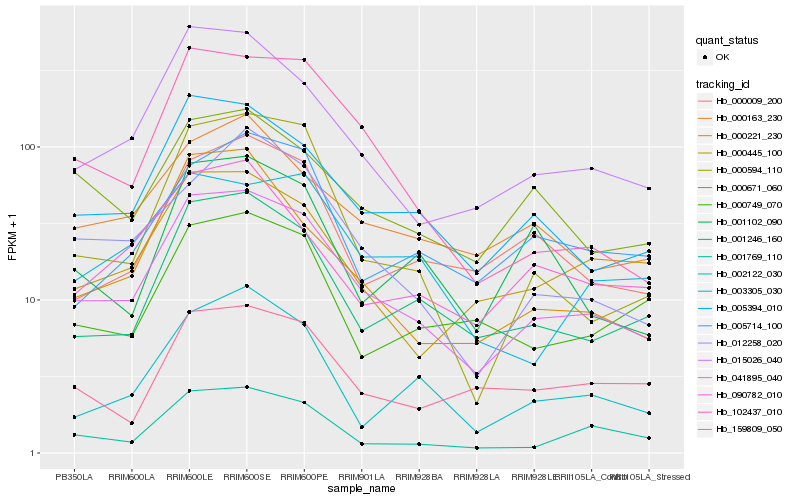
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_041895_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005394_010 |
0.0949169227 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 3 |
Hb_015026_040 |
0.101333454 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 4 |
Hb_001246_160 |
0.114869441 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 5 |
Hb_000163_230 |
0.1307601449 |
- |
- |
Phytochrome A-associated F-box protein, putative [Ricinus communis] |
| 6 |
Hb_001769_110 |
0.1330532626 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 7 |
Hb_000594_110 |
0.1353900441 |
transcription factor |
TF Family: RAV |
DNA-binding protein RAV1, putative [Ricinus communis] |
| 8 |
Hb_000009_200 |
0.1357430883 |
- |
- |
PREDICTED: UPF0496 protein At2g18630 [Jatropha curcas] |
| 9 |
Hb_102437_010 |
0.1366254884 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_000445_100 |
0.1367934125 |
- |
- |
PREDICTED: glutathione S-transferase TCHQD [Jatropha curcas] |
| 11 |
Hb_012258_020 |
0.1370597074 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 12 |
Hb_000671_060 |
0.1389030271 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_000221_230 |
0.1389884655 |
transcription factor |
TF Family: Jumonji |
PREDICTED: F-box protein At1g78280 [Jatropha curcas] |
| 14 |
Hb_003305_030 |
0.143727672 |
- |
- |
- |
| 15 |
Hb_002122_030 |
0.1449606688 |
- |
- |
- |
| 16 |
Hb_090782_010 |
0.1464938194 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 17 |
Hb_005714_100 |
0.1472333479 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |
| 18 |
Hb_000749_070 |
0.1493329513 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 19 |
Hb_159809_050 |
0.1500734769 |
- |
- |
AT14A, putative [Ricinus communis] |
| 20 |
Hb_001102_090 |
0.1535364321 |
- |
- |
PREDICTED: mitogen-activated protein kinase 19 [Jatropha curcas] |