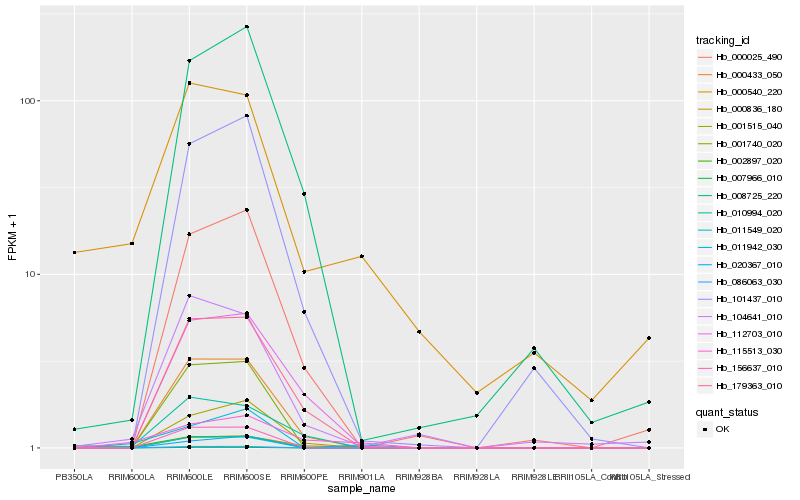
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002897_020 |
0.0 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 2 |
Hb_115513_030 |
0.1692340669 |
- |
- |
PREDICTED: uncharacterized protein At2g34160 [Jatropha curcas] |
| 3 |
Hb_011942_030 |
0.1759565266 |
- |
- |
- |
| 4 |
Hb_000836_180 |
0.177127891 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 5 |
Hb_000433_050 |
0.1942356152 |
- |
- |
PREDICTED: cell number regulator 1 isoform X1 [Vitis vinifera] |
| 6 |
Hb_104641_010 |
0.2036119045 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_179363_010 |
0.2086461214 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 8 |
Hb_000540_220 |
0.2138648474 |
- |
- |
hypothetical protein JCGZ_05605 [Jatropha curcas] |
| 9 |
Hb_101437_010 |
0.2176489413 |
- |
- |
PREDICTED: protein P21-like [Jatropha curcas] |
| 10 |
Hb_001515_040 |
0.2183642356 |
- |
- |
- |
| 11 |
Hb_010994_020 |
0.2202921254 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_112703_010 |
0.2220922527 |
- |
- |
PREDICTED: cytochrome P450 CYP72A219-like [Prunus mume] |
| 13 |
Hb_000025_490 |
0.2233251452 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 14 |
Hb_020367_010 |
0.2236509484 |
- |
- |
hypothetical protein PRUPE_ppa004271mg [Prunus persica] |
| 15 |
Hb_086063_030 |
0.2249000255 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8-like [Camelina sativa] |
| 16 |
Hb_007966_010 |
0.2249098898 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 17 |
Hb_011549_020 |
0.2249351546 |
- |
- |
- |
| 18 |
Hb_156637_010 |
0.2249614505 |
- |
- |
PREDICTED: aldose reductase-like [Camelina sativa] |
| 19 |
Hb_001740_020 |
0.2249826969 |
- |
- |
hypothetical protein JCGZ_23765 [Jatropha curcas] |
| 20 |
Hb_008725_220 |
0.2250048861 |
- |
- |
PREDICTED: uncharacterized protein LOC105642593 [Jatropha curcas] |