| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
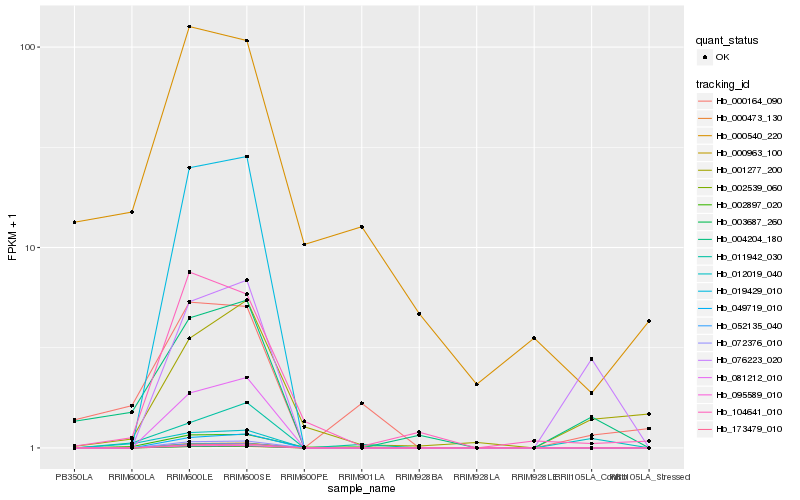
Hb_004204_180 |
0.0 |
- |
- |
- |
| 2 |
Hb_000164_090 |
0.2493709064 |
- |
- |
hypothetical protein [Plasmodium berghei strain ANKA], partial [Plasmodium berghei ANKA] |
| 3 |
Hb_012019_040 |
0.2652304914 |
- |
- |
- |
| 4 |
Hb_000540_220 |
0.2675131908 |
- |
- |
hypothetical protein JCGZ_05605 [Jatropha curcas] |
| 5 |
Hb_076223_020 |
0.2743669643 |
- |
- |
- |
| 6 |
Hb_104641_010 |
0.2771292928 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_011942_030 |
0.278243668 |
- |
- |
- |
| 8 |
Hb_002897_020 |
0.284481848 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 9 |
Hb_173479_010 |
0.286657214 |
- |
- |
- |
| 10 |
Hb_003687_260 |
0.2867058443 |
- |
- |
PREDICTED: inorganic phosphate transporter 1-4-like [Camelina sativa] |
| 11 |
Hb_002539_060 |
0.2867301613 |
- |
- |
- |
| 12 |
Hb_049719_010 |
0.2868642217 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_081212_010 |
0.287238704 |
- |
- |
- |
| 14 |
Hb_001277_200 |
0.2873211731 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 15 |
Hb_019429_010 |
0.2876936812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_072376_010 |
0.2881497801 |
- |
- |
- |
| 17 |
Hb_000963_100 |
0.2881774772 |
- |
- |
Protein SUA5, putative [Ricinus communis] |
| 18 |
Hb_052135_040 |
0.2881836108 |
- |
- |
- |
| 19 |
Hb_095589_010 |
0.288210414 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 20 |
Hb_000473_130 |
0.2882229645 |
- |
- |
PREDICTED: uncharacterized protein LOC105778857 [Gossypium raimondii] |