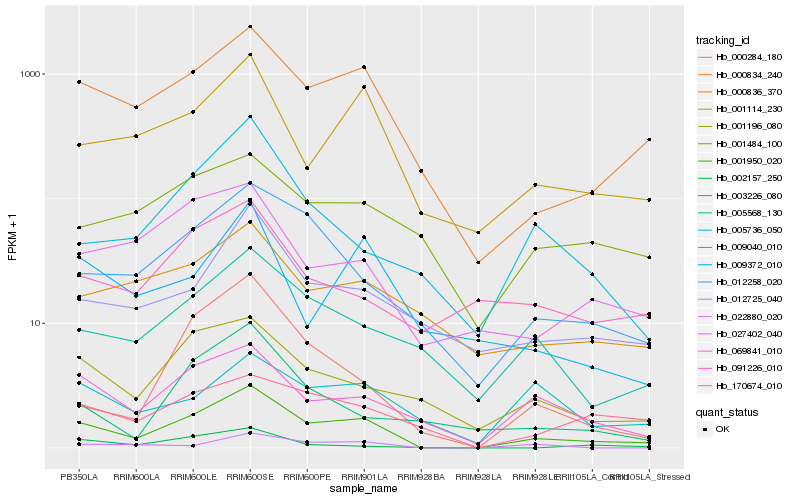
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001950_020 |
0.0 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 2 |
Hb_000834_240 |
0.1736974909 |
- |
- |
hypothetical protein JCGZ_07230 [Jatropha curcas] |
| 3 |
Hb_069841_010 |
0.2051520169 |
- |
- |
resistance protein [Quercus suber] |
| 4 |
Hb_001114_230 |
0.2061985753 |
- |
- |
- |
| 5 |
Hb_012258_020 |
0.2117417998 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 6 |
Hb_001196_080 |
0.2176246721 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 7 |
Hb_005568_130 |
0.2182349724 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_012725_040 |
0.227179494 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 9 |
Hb_027402_040 |
0.22978436 |
- |
- |
Thioredoxin II, putative [Ricinus communis] |
| 10 |
Hb_009040_010 |
0.2331826166 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_000284_180 |
0.2371635742 |
- |
- |
hypothetical protein EUGRSUZ_B01865 [Eucalyptus grandis] |
| 12 |
Hb_022880_020 |
0.2378636354 |
- |
- |
PREDICTED: uncharacterized protein LOC105631661 [Jatropha curcas] |
| 13 |
Hb_170674_010 |
0.239368414 |
- |
- |
hypothetical protein JCGZ_23765 [Jatropha curcas] |
| 14 |
Hb_000836_370 |
0.2427747038 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_009372_010 |
0.2431868786 |
- |
- |
- |
| 16 |
Hb_091226_010 |
0.249895703 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA isoform X1 [Jatropha curcas] |
| 17 |
Hb_005736_050 |
0.2504344261 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 18 |
Hb_002157_250 |
0.250831084 |
- |
- |
Pectinesterase-4 precursor, putative [Ricinus communis] |
| 19 |
Hb_003226_080 |
0.2512196034 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5-like [Jatropha curcas] |
| 20 |
Hb_001484_100 |
0.2517523061 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |