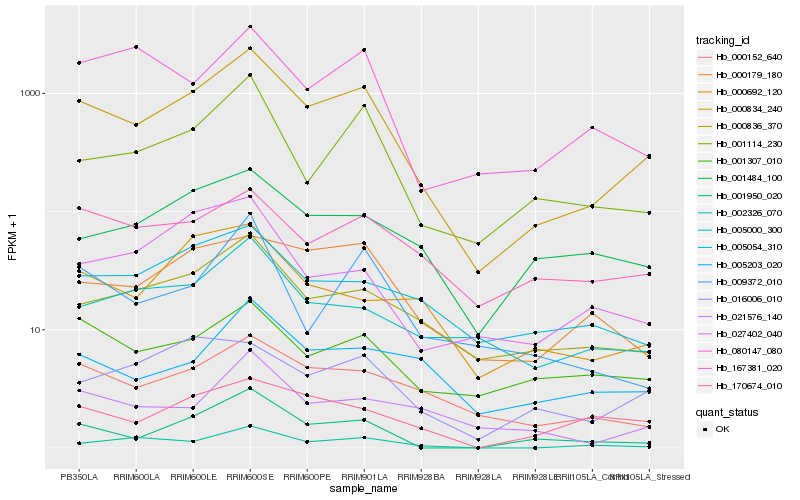
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000834_240 |
0.0 |
- |
- |
hypothetical protein JCGZ_07230 [Jatropha curcas] |
| 2 |
Hb_000836_370 |
0.1583068328 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_001114_230 |
0.1618661621 |
- |
- |
- |
| 4 |
Hb_000152_640 |
0.1672648226 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKS1 [Jatropha curcas] |
| 5 |
Hb_000179_180 |
0.1707077286 |
- |
- |
hypothetical protein EUGRSUZ_C031461, partial [Eucalyptus grandis] |
| 6 |
Hb_001950_020 |
0.1736974909 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 7 |
Hb_005054_310 |
0.1824630823 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 8 |
Hb_001307_010 |
0.1827293007 |
- |
- |
PREDICTED: uncharacterized protein LOC103939917 [Pyrus x bretschneideri] |
| 9 |
Hb_016006_010 |
0.1833640392 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 10 |
Hb_167381_020 |
0.1842520863 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 11 |
Hb_005203_020 |
0.1846948936 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 12 |
Hb_001484_100 |
0.185238778 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 13 |
Hb_170674_010 |
0.1857746733 |
- |
- |
hypothetical protein JCGZ_23765 [Jatropha curcas] |
| 14 |
Hb_005000_300 |
0.1871663628 |
transcription factor |
TF Family: TRAF |
putative regulatory protein NPR1 [Hevea brasiliensis] |
| 15 |
Hb_080147_080 |
0.1872581326 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 16 |
Hb_000692_120 |
0.1880922526 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 17 |
Hb_021576_140 |
0.1882579572 |
- |
- |
PREDICTED: protein POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION-like [Jatropha curcas] |
| 18 |
Hb_027402_040 |
0.188313075 |
- |
- |
Thioredoxin II, putative [Ricinus communis] |
| 19 |
Hb_002326_070 |
0.1926255622 |
- |
- |
ent-kaurene oxidase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_009372_010 |
0.1931200022 |
- |
- |
- |