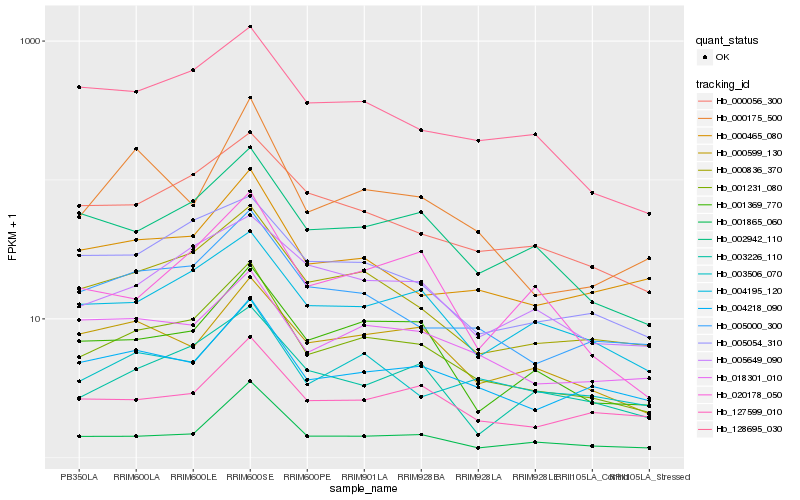
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001231_080 |
0.0 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like isoform X1 [Fragaria vesca subsp. vesca] |
| 2 |
Hb_000836_370 |
0.0860480105 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_005000_300 |
0.1008267597 |
transcription factor |
TF Family: TRAF |
putative regulatory protein NPR1 [Hevea brasiliensis] |
| 4 |
Hb_000056_300 |
0.1102302268 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002942_110 |
0.1108352675 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 2-like [Jatropha curcas] |
| 6 |
Hb_128695_030 |
0.1117407138 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 7 |
Hb_004195_120 |
0.1132033585 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 8 |
Hb_001369_770 |
0.1149763701 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 9 |
Hb_001865_060 |
0.1182476912 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 10 |
Hb_004218_090 |
0.1207813085 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_005054_310 |
0.1233921727 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 12 |
Hb_018301_010 |
0.1247121396 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 13 |
Hb_005649_090 |
0.1264123894 |
- |
- |
PREDICTED: ninja-family protein AFP3-like [Jatropha curcas] |
| 14 |
Hb_003506_070 |
0.1304095567 |
- |
- |
PREDICTED: calcineurin B-like protein 7 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000465_080 |
0.1309139773 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 16 |
Hb_003226_110 |
0.1319064253 |
- |
- |
PREDICTED: exocyst complex component SEC15B [Jatropha curcas] |
| 17 |
Hb_000599_130 |
0.1322903134 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 18 |
Hb_000175_500 |
0.133884037 |
- |
- |
PREDICTED: uncharacterized protein At1g15400 [Populus euphratica] |
| 19 |
Hb_127599_010 |
0.1346500261 |
- |
- |
hypothetical protein CISIN_1g0010462mg, partial [Citrus sinensis] |
| 20 |
Hb_020178_050 |
0.1366085809 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |