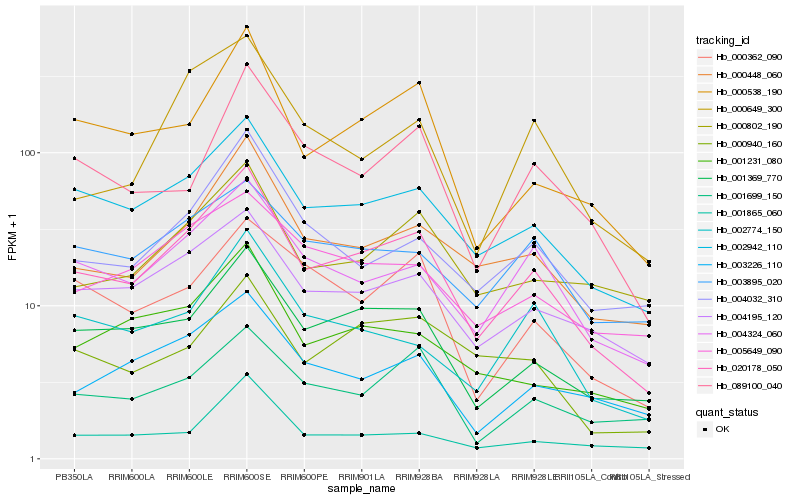
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020178_050 |
0.0 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 2 |
Hb_002942_110 |
0.0925110159 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 2-like [Jatropha curcas] |
| 3 |
Hb_001369_770 |
0.1030472491 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 4 |
Hb_000448_060 |
0.1130640665 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 5 |
Hb_004324_060 |
0.113628765 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 6 |
Hb_004195_120 |
0.1136523672 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 7 |
Hb_000538_190 |
0.1170265964 |
- |
- |
PREDICTED: protein DJ-1 homolog D [Jatropha curcas] |
| 8 |
Hb_003226_110 |
0.133731074 |
- |
- |
PREDICTED: exocyst complex component SEC15B [Jatropha curcas] |
| 9 |
Hb_004032_310 |
0.1360100337 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 10 |
Hb_001231_080 |
0.1366085809 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like isoform X1 [Fragaria vesca subsp. vesca] |
| 11 |
Hb_000802_190 |
0.1375218027 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 12 |
Hb_089100_040 |
0.1400075005 |
- |
- |
PREDICTED: uncharacterized protein LOC105638961 [Jatropha curcas] |
| 13 |
Hb_005649_090 |
0.1415592268 |
- |
- |
PREDICTED: ninja-family protein AFP3-like [Jatropha curcas] |
| 14 |
Hb_000362_090 |
0.1437916972 |
- |
- |
PREDICTED: uncharacterized protein LOC105635736 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002774_150 |
0.1441257362 |
- |
- |
PREDICTED: F-box protein At1g22220 [Jatropha curcas] |
| 16 |
Hb_000940_160 |
0.144651228 |
- |
- |
hypothetical protein VITISV_017596 [Vitis vinifera] |
| 17 |
Hb_001699_150 |
0.1451419207 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 18 |
Hb_001865_060 |
0.1462056233 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 19 |
Hb_003895_020 |
0.1462182071 |
- |
- |
PREDICTED: dedicator of cytokinesis protein 7 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000649_300 |
0.1467212688 |
transcription factor |
TF Family: Tify |
JAZ9 [Hevea brasiliensis] |