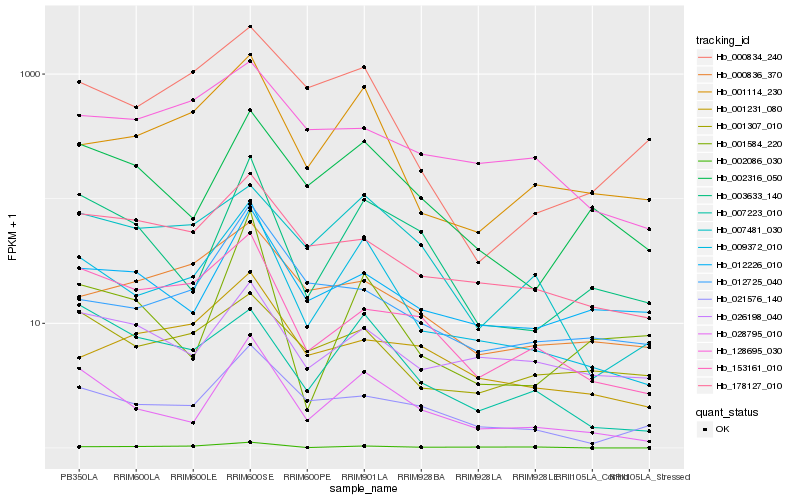
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009372_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_028795_010 |
0.1308656023 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 3 |
Hb_001114_230 |
0.1380510761 |
- |
- |
- |
| 4 |
Hb_021576_140 |
0.1845582606 |
- |
- |
PREDICTED: protein POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION-like [Jatropha curcas] |
| 5 |
Hb_007481_030 |
0.1886320057 |
- |
- |
PREDICTED: uncharacterized protein LOC105628433 [Jatropha curcas] |
| 6 |
Hb_153161_010 |
0.1893711083 |
- |
- |
phospholipid-transporting atpase, putative [Ricinus communis] |
| 7 |
Hb_003633_140 |
0.1905122278 |
- |
- |
PREDICTED: uncharacterized protein LOC105647486 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002316_050 |
0.1908280742 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007223_010 |
0.1928398418 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000834_240 |
0.1931200022 |
- |
- |
hypothetical protein JCGZ_07230 [Jatropha curcas] |
| 11 |
Hb_178127_010 |
0.1946797255 |
- |
- |
Ubiquitin ligase SINAT3, putative [Ricinus communis] |
| 12 |
Hb_002086_030 |
0.1966292575 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 13 |
Hb_000836_370 |
0.2063239566 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_012725_040 |
0.2087288824 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 15 |
Hb_001231_080 |
0.2105978357 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like isoform X1 [Fragaria vesca subsp. vesca] |
| 16 |
Hb_001307_010 |
0.2107041405 |
- |
- |
PREDICTED: uncharacterized protein LOC103939917 [Pyrus x bretschneideri] |
| 17 |
Hb_128695_030 |
0.2120895485 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 18 |
Hb_001584_220 |
0.2120901499 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 19 |
Hb_012226_010 |
0.2125372551 |
- |
- |
PREDICTED: protein DGCR14 [Jatropha curcas] |
| 20 |
Hb_026198_040 |
0.2129786869 |
- |
- |
PREDICTED: uncharacterized protein LOC105628901 [Jatropha curcas] |