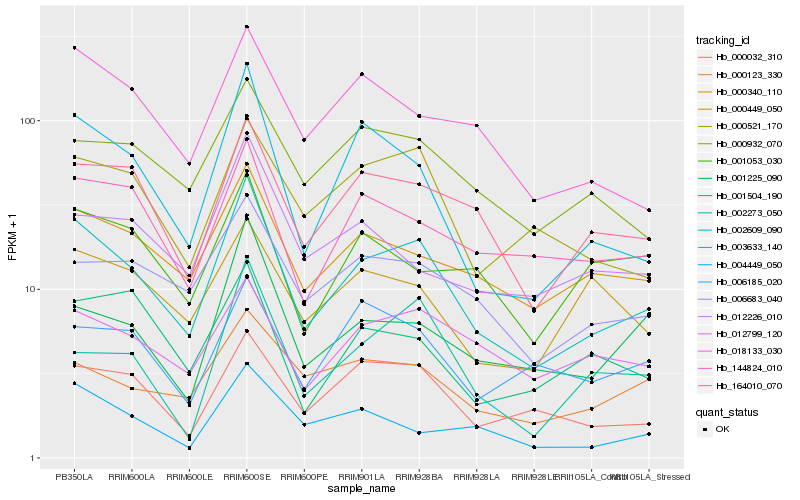
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002609_090 |
0.0 |
- |
- |
PREDICTED: phosphomethylpyrimidine synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 2 |
Hb_003633_140 |
0.1255523563 |
- |
- |
PREDICTED: uncharacterized protein LOC105647486 isoform X1 [Jatropha curcas] |
| 3 |
Hb_164010_070 |
0.1329231233 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 4 |
Hb_000340_110 |
0.1368261721 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X1 [Jatropha curcas] |
| 5 |
Hb_000032_310 |
0.1395116701 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 6 |
Hb_144824_010 |
0.1475995068 |
- |
- |
hypothetical protein POPTR_0019s02110g [Populus trichocarpa] |
| 7 |
Hb_006683_040 |
0.1477194443 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 8 |
Hb_000521_170 |
0.1486629563 |
- |
- |
PREDICTED: callose synthase 1 [Jatropha curcas] |
| 9 |
Hb_018133_030 |
0.149596798 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 10 |
Hb_001225_090 |
0.1554009069 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_000932_070 |
0.1560595706 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 12 |
Hb_000123_330 |
0.1566848387 |
- |
- |
hypothetical protein M569_14136, partial [Genlisea aurea] |
| 13 |
Hb_012799_120 |
0.1569449926 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004449_050 |
0.1618558248 |
- |
- |
PREDICTED: tyrosine-sulfated glycopeptide receptor 1-like [Jatropha curcas] |
| 15 |
Hb_012226_010 |
0.1659329621 |
- |
- |
PREDICTED: protein DGCR14 [Jatropha curcas] |
| 16 |
Hb_001504_190 |
0.1666661858 |
- |
- |
double-stranded RNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_002273_050 |
0.1666824738 |
- |
- |
PREDICTED: geranylgeranyl diphosphate reductase, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_001053_030 |
0.1675745699 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_006185_020 |
0.1686168039 |
- |
- |
PREDICTED: midasin [Jatropha curcas] |
| 20 |
Hb_000449_050 |
0.1687248068 |
- |
- |
Protein bem46, putative [Ricinus communis] |