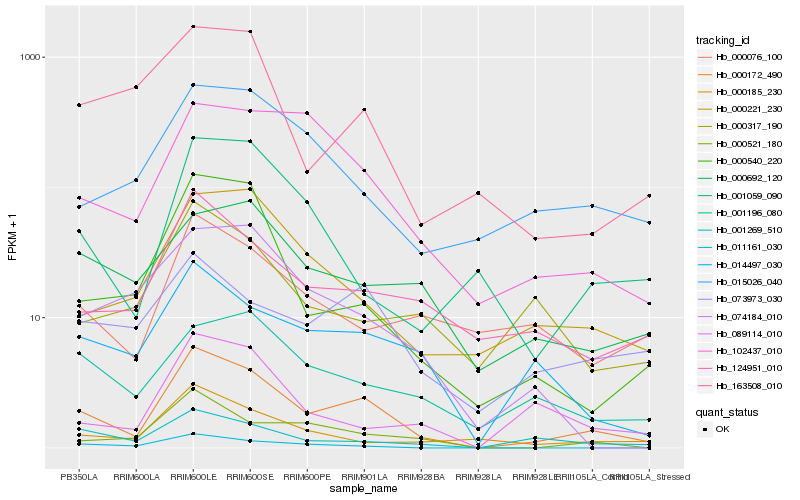
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000172_490 |
0.0 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 2 |
Hb_014497_030 |
0.2161664976 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein BUBR1 [Jatropha curcas] |
| 3 |
Hb_000540_220 |
0.2183998027 |
- |
- |
hypothetical protein JCGZ_05605 [Jatropha curcas] |
| 4 |
Hb_124951_010 |
0.2193322061 |
- |
- |
chitinase-like protein [Hevea brasiliensis] |
| 5 |
Hb_000221_230 |
0.2225116819 |
transcription factor |
TF Family: Jumonji |
PREDICTED: F-box protein At1g78280 [Jatropha curcas] |
| 6 |
Hb_000521_180 |
0.2262325166 |
- |
- |
PREDICTED: uncharacterized protein LOC105650335 [Jatropha curcas] |
| 7 |
Hb_001269_510 |
0.226789242 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 8 |
Hb_000185_230 |
0.2330448844 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 9 |
Hb_102437_010 |
0.2362389044 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_089114_010 |
0.2380613724 |
- |
- |
PREDICTED: uncharacterized protein LOC105646926 [Jatropha curcas] |
| 11 |
Hb_073973_030 |
0.2391430495 |
- |
- |
PREDICTED: uncharacterized protein LOC105638905 isoform X2 [Jatropha curcas] |
| 12 |
Hb_015026_040 |
0.2396787989 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 13 |
Hb_001059_090 |
0.2408362501 |
- |
- |
PREDICTED: F-box protein At1g61340 [Jatropha curcas] |
| 14 |
Hb_011161_030 |
0.2408912817 |
- |
- |
PREDICTED: uncharacterized protein LOC105638906 [Jatropha curcas] |
| 15 |
Hb_163508_010 |
0.2417996489 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 16 |
Hb_001196_080 |
0.2451971234 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 17 |
Hb_000317_190 |
0.2498661083 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 18 |
Hb_000076_100 |
0.2499411515 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1 [Jatropha curcas] |
| 19 |
Hb_074184_010 |
0.2502375547 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 20 |
Hb_000692_120 |
0.250526749 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |