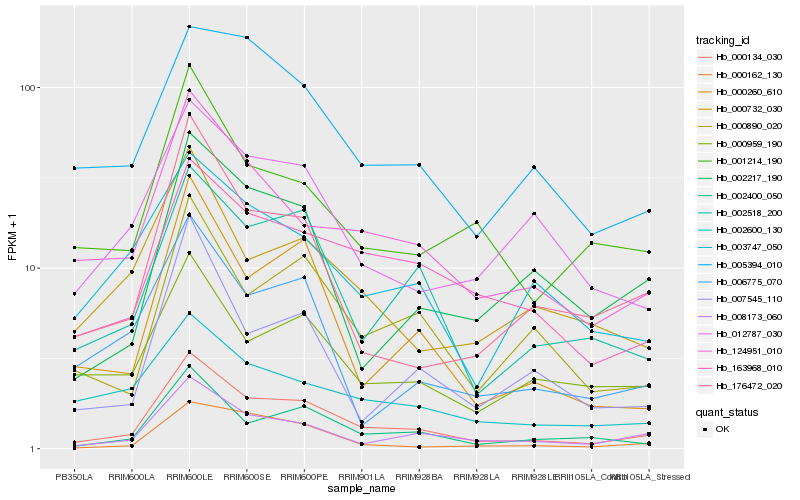
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000134_030 |
0.0 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 3-like [Jatropha curcas] |
| 2 |
Hb_008173_060 |
0.1288140082 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 3 |
Hb_002400_050 |
0.1352721225 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X1 [Jatropha curcas] |
| 4 |
Hb_124951_010 |
0.1386182101 |
- |
- |
chitinase-like protein [Hevea brasiliensis] |
| 5 |
Hb_002518_200 |
0.1401492172 |
- |
- |
Proliferating cell nuclear antigen [Theobroma cacao] |
| 6 |
Hb_000260_610 |
0.1468232147 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 7 |
Hb_000890_020 |
0.1518700235 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 8 |
Hb_163968_010 |
0.1528250523 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 9 |
Hb_176472_020 |
0.1613842733 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001214_190 |
0.1680515997 |
- |
- |
hypothetical protein JCGZ_25001 [Jatropha curcas] |
| 11 |
Hb_000959_190 |
0.1693021002 |
- |
- |
PREDICTED: crossover junction endonuclease EME1B-like [Jatropha curcas] |
| 12 |
Hb_002217_190 |
0.1730956562 |
- |
- |
invertase [Hevea brasiliensis] |
| 13 |
Hb_007545_110 |
0.1740093119 |
- |
- |
catalytic, putative [Ricinus communis] |
| 14 |
Hb_006775_070 |
0.1743776521 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_005394_010 |
0.1743810373 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 16 |
Hb_000732_030 |
0.1751437382 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_012787_030 |
0.1766101022 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 18 |
Hb_002600_130 |
0.1768092207 |
- |
- |
PREDICTED: uncharacterized protein LOC105646808 [Jatropha curcas] |
| 19 |
Hb_003747_050 |
0.1771313172 |
- |
- |
PREDICTED: WD repeat-containing protein LWD1 [Jatropha curcas] |
| 20 |
Hb_000162_130 |
0.1775803962 |
- |
- |
conserved hypothetical protein [Ricinus communis] |