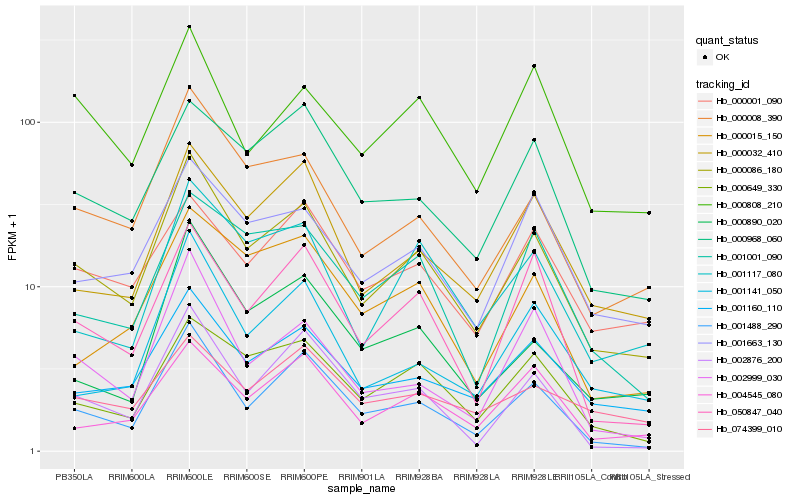
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000086_180 |
0.0 |
- |
- |
hypothetical protein JCGZ_17674 [Jatropha curcas] |
| 2 |
Hb_000008_390 |
0.0868492636 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 3 |
Hb_001488_290 |
0.0996677752 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 4 |
Hb_001160_110 |
0.1056388217 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 5 |
Hb_000032_410 |
0.1198391768 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 6 |
Hb_001663_130 |
0.123170452 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 7 |
Hb_000649_330 |
0.1233877404 |
- |
- |
hypothetical protein POPTR_0010s11880g [Populus trichocarpa] |
| 8 |
Hb_001117_080 |
0.1250190003 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_050847_040 |
0.1277645049 |
- |
- |
PREDICTED: uncharacterized protein LOC105649141 [Jatropha curcas] |
| 10 |
Hb_000968_060 |
0.1315922197 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 11 |
Hb_000890_020 |
0.1337074619 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 12 |
Hb_004545_080 |
0.1342563656 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_001141_050 |
0.1351221267 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000808_210 |
0.1366098927 |
- |
- |
hypothetical protein EUTSA_v10023481mg [Eutrema salsugineum] |
| 15 |
Hb_002999_030 |
0.1387375532 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 16 |
Hb_002876_200 |
0.139005347 |
- |
- |
PREDICTED: uncharacterized protein LOC105641209 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000001_090 |
0.1390969304 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 18 |
Hb_001001_090 |
0.139869325 |
- |
- |
PREDICTED: uncharacterized protein LOC105633658 [Jatropha curcas] |
| 19 |
Hb_000015_150 |
0.1411381756 |
- |
- |
Ubiquitin-conjugating enzyme E2 7 family protein [Populus trichocarpa] |
| 20 |
Hb_074399_010 |
0.1416260818 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |