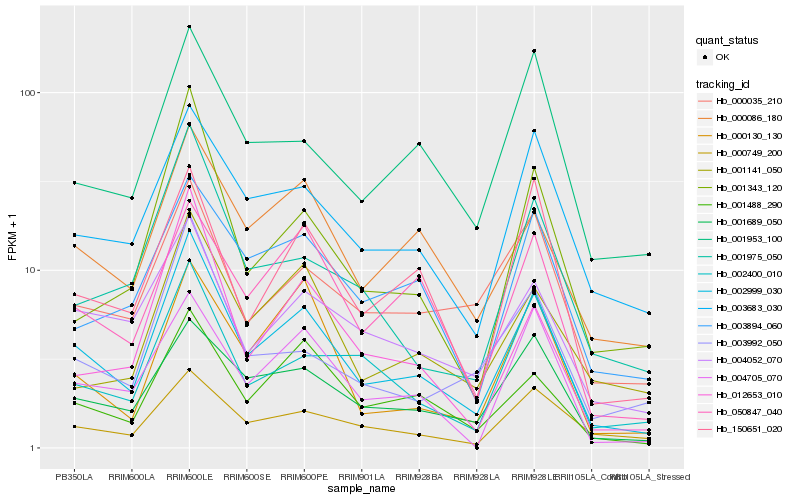
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002999_030 |
0.0 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 2 |
Hb_001488_290 |
0.1302145809 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 3 |
Hb_001689_050 |
0.136354282 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Prunus mume] |
| 4 |
Hb_000086_180 |
0.1387375532 |
- |
- |
hypothetical protein JCGZ_17674 [Jatropha curcas] |
| 5 |
Hb_004052_070 |
0.1406273198 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 6 |
Hb_003683_030 |
0.1445136548 |
- |
- |
PREDICTED: uridine-cytidine kinase C isoform X1 [Jatropha curcas] |
| 7 |
Hb_000749_200 |
0.1511664857 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001141_050 |
0.1529923133 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001975_050 |
0.1530284238 |
- |
- |
Tyrosine-specific transport protein, putative [Ricinus communis] |
| 10 |
Hb_001343_120 |
0.1543509106 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_150651_020 |
0.1555532937 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 12 |
Hb_004705_070 |
0.1557022676 |
- |
- |
PREDICTED: uncharacterized protein LOC105647312 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000035_210 |
0.155784615 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |
| 14 |
Hb_012653_010 |
0.1557945582 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 15 |
Hb_050847_040 |
0.1567742254 |
- |
- |
PREDICTED: uncharacterized protein LOC105649141 [Jatropha curcas] |
| 16 |
Hb_003894_060 |
0.1576744699 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003992_050 |
0.1581363721 |
- |
- |
delta 9 desaturase, putative [Ricinus communis] |
| 18 |
Hb_002400_010 |
0.1583730508 |
- |
- |
PREDICTED: putative L-ascorbate peroxidase 6 [Jatropha curcas] |
| 19 |
Hb_000130_130 |
0.1598085085 |
- |
- |
hypothetical protein POPTR_0001s40330g [Populus trichocarpa] |
| 20 |
Hb_001953_100 |
0.1599399352 |
- |
- |
PREDICTED: glutamate-1-semialdehyde 2,1-aminomutase 2, chloroplastic [Jatropha curcas] |