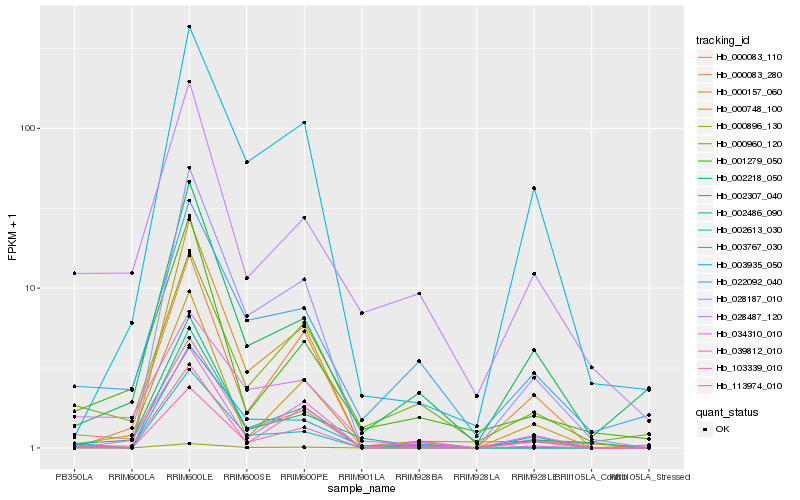
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000083_110 |
0.0 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 2 |
Hb_103339_010 |
0.1553834025 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_001279_050 |
0.1693720616 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 4 |
Hb_002307_040 |
0.1721992759 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_028487_120 |
0.1754557541 |
- |
- |
PREDICTED: HMG-Y-related protein B-like [Jatropha curcas] |
| 6 |
Hb_000960_120 |
0.1763458814 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 7 |
Hb_002613_030 |
0.1787060124 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000083_280 |
0.1796997212 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 9 |
Hb_039812_010 |
0.1801069844 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 10 |
Hb_113974_010 |
0.1806404799 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_002486_090 |
0.1810820898 |
- |
- |
hypothetical protein POPTR_0005s06910g [Populus trichocarpa] |
| 12 |
Hb_000896_130 |
0.1814271436 |
transcription factor |
TF Family: CPP |
cysteine-rich polycomb-like family protein [Populus trichocarpa] |
| 13 |
Hb_034310_010 |
0.1818072685 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_000748_100 |
0.1837929644 |
- |
- |
hypothetical protein POPTR_0002s06160g [Populus trichocarpa] |
| 15 |
Hb_003935_050 |
0.1862590261 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_028187_010 |
0.1882483828 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 17 |
Hb_002218_050 |
0.1884366638 |
- |
- |
SSXT family protein isoform 2, partial [Theobroma cacao] |
| 18 |
Hb_022092_040 |
0.1886441779 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 [Jatropha curcas] |
| 19 |
Hb_000157_060 |
0.1890685418 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003767_030 |
0.1949667302 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |