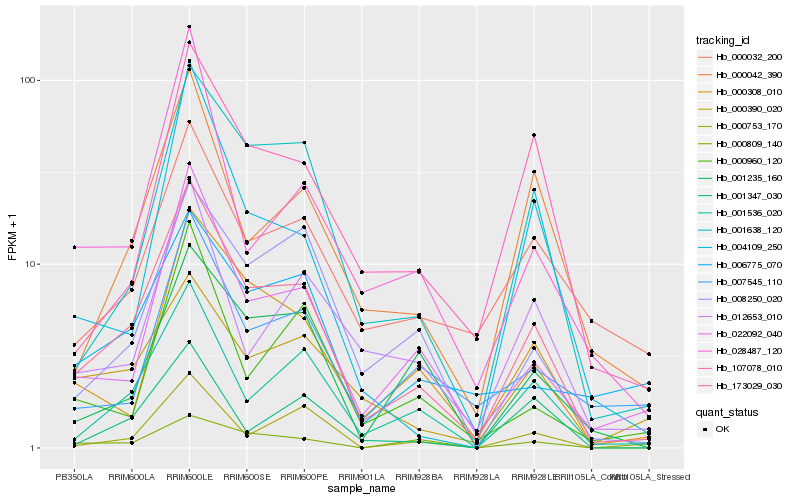
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_173029_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001536_020 |
0.1488268403 |
transcription factor |
TF Family: GRF |
PREDICTED: uncharacterized protein LOC105631006 [Jatropha curcas] |
| 3 |
Hb_001638_120 |
0.1506325707 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 4 |
Hb_022092_040 |
0.1533788643 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 [Jatropha curcas] |
| 5 |
Hb_000308_010 |
0.1609090605 |
- |
- |
PREDICTED: uncharacterized protein LOC105639684 [Jatropha curcas] |
| 6 |
Hb_000042_390 |
0.1748667932 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7 [Jatropha curcas] |
| 7 |
Hb_008250_020 |
0.1829130454 |
- |
- |
PREDICTED: U-box domain-containing protein 15-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001235_160 |
0.1839136843 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_107078_010 |
0.185202163 |
- |
- |
- |
| 10 |
Hb_000390_020 |
0.1858783056 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 4-like [Jatropha curcas] |
| 11 |
Hb_000809_140 |
0.1901469976 |
- |
- |
hypothetical protein JCGZ_12354 [Jatropha curcas] |
| 12 |
Hb_012653_010 |
0.1914026775 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 13 |
Hb_000032_200 |
0.1920709923 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 14 |
Hb_007545_110 |
0.1933733811 |
- |
- |
catalytic, putative [Ricinus communis] |
| 15 |
Hb_000753_170 |
0.1942656818 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_028487_120 |
0.1962264848 |
- |
- |
PREDICTED: HMG-Y-related protein B-like [Jatropha curcas] |
| 17 |
Hb_006775_070 |
0.2006848827 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_000960_120 |
0.2017521119 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 19 |
Hb_001347_030 |
0.2019943042 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_004109_250 |
0.2020135459 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |