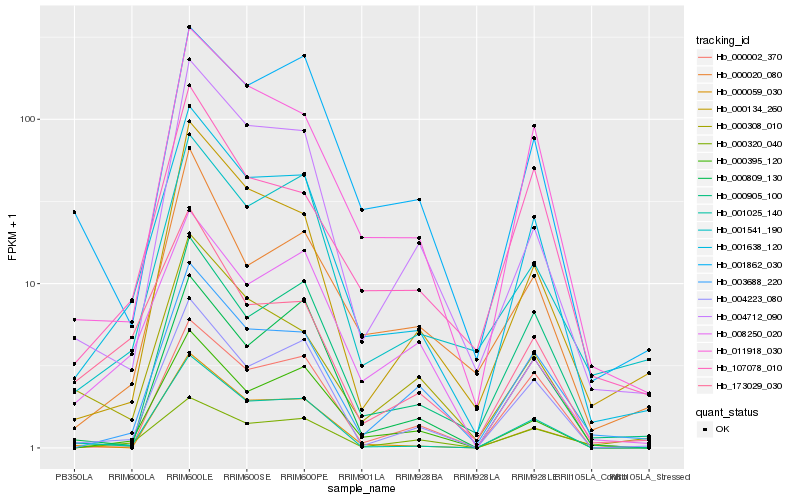
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001638_120 |
0.0 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 2 |
Hb_011918_030 |
0.1011775987 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 3 |
Hb_004223_080 |
0.118180554 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 4 |
Hb_001025_140 |
0.1213413715 |
- |
- |
hypothetical protein POPTR_0015s09180g [Populus trichocarpa] |
| 5 |
Hb_003688_220 |
0.1339375747 |
- |
- |
unknown [Populus trichocarpa] |
| 6 |
Hb_000134_260 |
0.1359556415 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_107078_010 |
0.1360889782 |
- |
- |
- |
| 8 |
Hb_001541_190 |
0.1387878885 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 9 |
Hb_004712_090 |
0.1388395073 |
- |
- |
Serine acetyltransferase 3, mitochondrial precursor, putative [Ricinus communis] |
| 10 |
Hb_000395_120 |
0.1399749524 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 11 |
Hb_000320_040 |
0.1433580607 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_000905_100 |
0.1501941522 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_173029_030 |
0.1506325707 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000308_010 |
0.1523243307 |
- |
- |
PREDICTED: uncharacterized protein LOC105639684 [Jatropha curcas] |
| 15 |
Hb_000059_030 |
0.1550357457 |
- |
- |
PREDICTED: chaperone protein dnaJ 10 [Jatropha curcas] |
| 16 |
Hb_000020_080 |
0.1553425834 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_09080 [Jatropha curcas] |
| 17 |
Hb_001862_030 |
0.1562738654 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 18 |
Hb_008250_020 |
0.1599175366 |
- |
- |
PREDICTED: U-box domain-containing protein 15-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000002_370 |
0.160331492 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 20 |
Hb_000809_130 |
0.1615281294 |
- |
- |
PREDICTED: peroxidase 10 isoform X3 [Jatropha curcas] |