| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
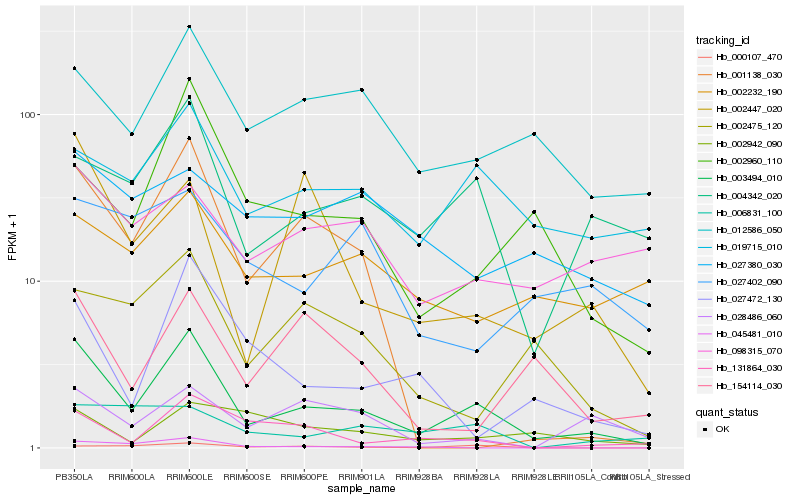
Hb_003494_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_19838 [Jatropha curcas] |
| 2 |
Hb_004342_020 |
0.2323450265 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_012586_050 |
0.2449550436 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Prunus mume] |
| 4 |
Hb_002960_110 |
0.2467777191 |
- |
- |
PREDICTED: synaptotagmin-2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_019715_010 |
0.2488994137 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 6 |
Hb_002447_020 |
0.2605766229 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_027472_130 |
0.263604156 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 8 |
Hb_131864_030 |
0.2652928486 |
transcription factor |
TF Family: C2H2 |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_001138_030 |
0.2691445887 |
- |
- |
STRESS ENHANCED protein 1 [Populus trichocarpa] |
| 10 |
Hb_002475_120 |
0.27295142 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 11 |
Hb_154114_030 |
0.2761528809 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_002232_190 |
0.2795462747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_098315_070 |
0.2812559744 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105636030 isoform X1 [Jatropha curcas] |
| 14 |
Hb_028486_060 |
0.2819406287 |
- |
- |
PREDICTED: putative germin-like protein 9-2 [Jatropha curcas] |
| 15 |
Hb_045481_010 |
0.2842016466 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000107_470 |
0.2860500247 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 17 |
Hb_027380_030 |
0.29086961 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 18 |
Hb_002942_090 |
0.2938784384 |
- |
- |
DNAJ heat shock N-terminal domain-containing protein, putative [Theobroma cacao] |
| 19 |
Hb_006831_100 |
0.2940589324 |
- |
- |
snRNA-activating protein complex subunit, putative [Ricinus communis] |
| 20 |
Hb_027402_090 |
0.2952674239 |
- |
- |
endonuclease, putative [Ricinus communis] |