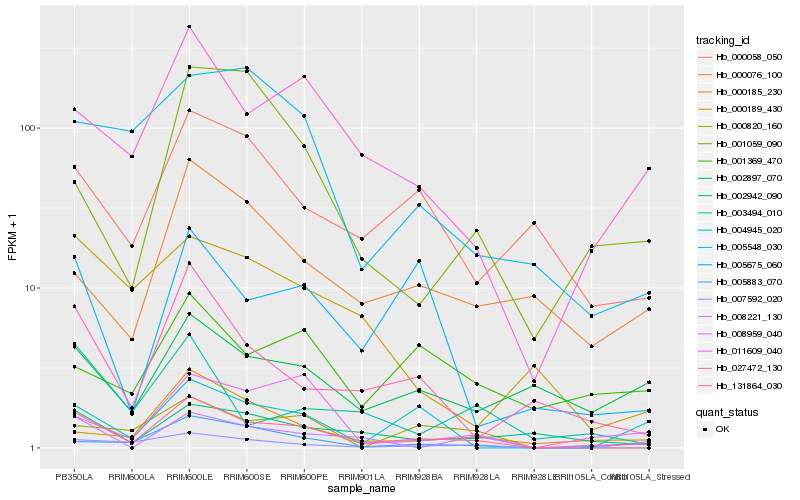
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_131864_030 |
0.0 |
transcription factor |
TF Family: C2H2 |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_011609_040 |
0.2008497947 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 40 [Jatropha curcas] |
| 3 |
Hb_027472_130 |
0.2205978061 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 4 |
Hb_005883_070 |
0.2325171499 |
- |
- |
PREDICTED: probable myosin-binding protein 5 [Jatropha curcas] |
| 5 |
Hb_002942_090 |
0.2371801778 |
- |
- |
DNAJ heat shock N-terminal domain-containing protein, putative [Theobroma cacao] |
| 6 |
Hb_000058_050 |
0.2415242102 |
- |
- |
PREDICTED: uncharacterized protein LOC105640877 [Jatropha curcas] |
| 7 |
Hb_005675_060 |
0.242906293 |
- |
- |
PREDICTED: protein NIM1-INTERACTING 2 [Jatropha curcas] |
| 8 |
Hb_002897_070 |
0.2455080014 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 9 |
Hb_008959_040 |
0.2482859434 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 10 |
Hb_000185_230 |
0.2482886994 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 11 |
Hb_000076_100 |
0.2548470618 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1 [Jatropha curcas] |
| 12 |
Hb_005548_030 |
0.2575018496 |
- |
- |
hypothetical protein JCGZ_05606 [Jatropha curcas] |
| 13 |
Hb_004945_020 |
0.2587402538 |
- |
- |
PREDICTED: probable calcium-binding protein CML30 [Vitis vinifera] |
| 14 |
Hb_007592_020 |
0.2648058022 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 15 |
Hb_003494_010 |
0.2652928486 |
- |
- |
hypothetical protein JCGZ_19838 [Jatropha curcas] |
| 16 |
Hb_001369_470 |
0.2673836392 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKS1 [Jatropha curcas] |
| 17 |
Hb_001059_090 |
0.2686457178 |
- |
- |
PREDICTED: F-box protein At1g61340 [Jatropha curcas] |
| 18 |
Hb_008221_130 |
0.2697533355 |
- |
- |
hypothetical protein VITISV_016971 [Vitis vinifera] |
| 19 |
Hb_000820_160 |
0.2701829875 |
- |
- |
- |
| 20 |
Hb_000189_430 |
0.2707547667 |
- |
- |
unnamed protein product [Vitis vinifera] |