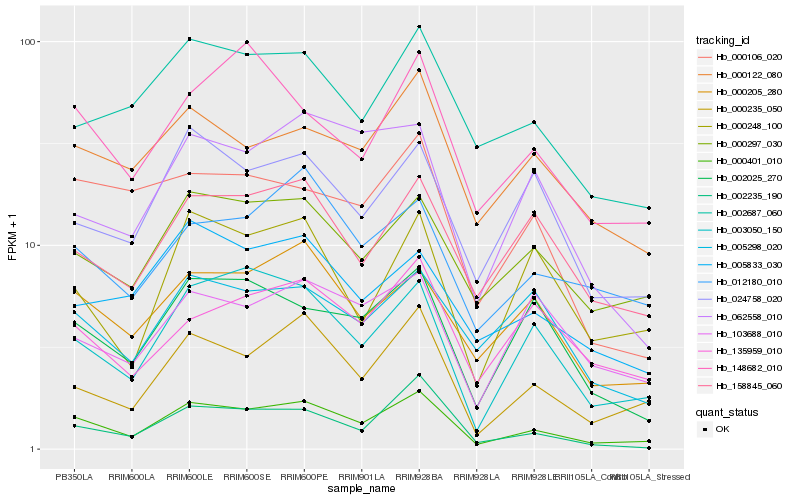
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000401_010 |
0.0 |
- |
- |
SAB, putative [Ricinus communis] |
| 2 |
Hb_000235_050 |
0.1005214817 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003050_150 |
0.1166108144 |
- |
- |
endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase, putative [Ricinus communis] |
| 4 |
Hb_002235_190 |
0.133581194 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000205_280 |
0.1353260045 |
- |
- |
PREDICTED: inositol phosphorylceramide glucuronosyltransferase 1 [Jatropha curcas] |
| 6 |
Hb_000297_030 |
0.1370054566 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002025_270 |
0.1376428505 |
- |
- |
PREDICTED: uncharacterized protein At4g15970 isoform X1 [Jatropha curcas] |
| 8 |
Hb_024758_020 |
0.1415115203 |
- |
- |
acyl-CoA binding protein 3B [Vernicia fordii] |
| 9 |
Hb_002687_060 |
0.1436025582 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 10 |
Hb_158845_060 |
0.1445340804 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000122_080 |
0.145062528 |
- |
- |
PREDICTED: staphylococcal-like nuclease CAN2 [Jatropha curcas] |
| 12 |
Hb_000248_100 |
0.1455847938 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 13 |
Hb_012180_010 |
0.1462506225 |
- |
- |
PREDICTED: (S)-ureidoglycine aminohydrolase [Jatropha curcas] |
| 14 |
Hb_148682_010 |
0.1497946455 |
- |
- |
PREDICTED: uncharacterized protein LOC105630720 [Jatropha curcas] |
| 15 |
Hb_062558_010 |
0.1514548938 |
- |
- |
PREDICTED: L-2-hydroxyglutarate dehydrogenase, mitochondrial isoform X1 [Populus euphratica] |
| 16 |
Hb_135959_010 |
0.1541089369 |
- |
- |
hypothetical protein JCGZ_07060 [Jatropha curcas] |
| 17 |
Hb_005833_030 |
0.1544032965 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860 [Jatropha curcas] |
| 18 |
Hb_000106_020 |
0.1555734111 |
- |
- |
PREDICTED: protein TOPLESS isoform X1 [Jatropha curcas] |
| 19 |
Hb_103688_010 |
0.1562472052 |
- |
- |
PREDICTED: sucrose-phosphatase 2-like [Jatropha curcas] |
| 20 |
Hb_005298_020 |
0.1567282259 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 2 [Jatropha curcas] |