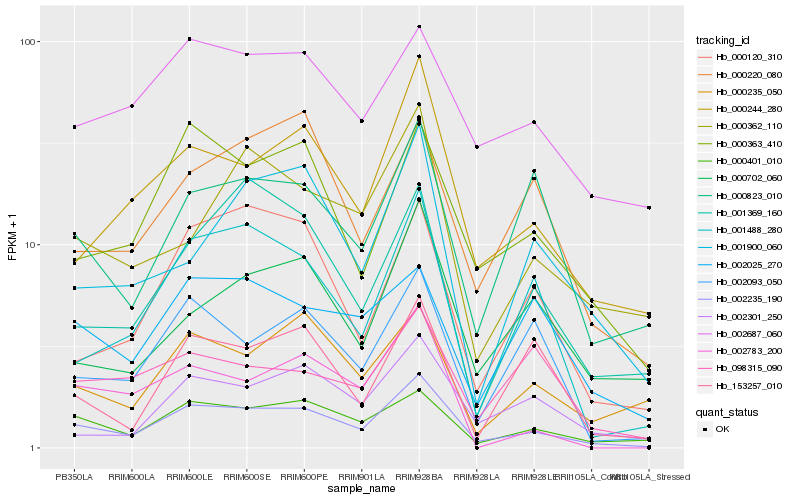
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002235_190 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000401_010 |
0.133581194 |
- |
- |
SAB, putative [Ricinus communis] |
| 3 |
Hb_000244_280 |
0.1356377949 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000363_410 |
0.1512645862 |
- |
- |
hypothetical protein JCGZ_00955 [Jatropha curcas] |
| 5 |
Hb_001488_280 |
0.1554175459 |
- |
- |
hypothetical protein JCGZ_19947 [Jatropha curcas] |
| 6 |
Hb_002093_050 |
0.1603882059 |
- |
- |
hypothetical protein POPTR_0008s02940g [Populus trichocarpa] |
| 7 |
Hb_000823_010 |
0.1685656834 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 8 |
Hb_000120_310 |
0.1690667461 |
- |
- |
Gibberellin 3-beta-dioxygenase, putative [Ricinus communis] |
| 9 |
Hb_002301_250 |
0.1711584481 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a [Jatropha curcas] |
| 10 |
Hb_000362_110 |
0.1719429117 |
- |
- |
PREDICTED: uncharacterized protein LOC104612097 isoform X2 [Nelumbo nucifera] |
| 11 |
Hb_000235_050 |
0.1720777482 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002687_060 |
0.1778622709 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 13 |
Hb_001369_160 |
0.1786272942 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 14 |
Hb_000702_060 |
0.1787986835 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001900_060 |
0.1791903699 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 16 |
Hb_000220_080 |
0.1793798576 |
- |
- |
kinase, putative [Ricinus communis] |
| 17 |
Hb_002783_200 |
0.1795900111 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 18 |
Hb_153257_010 |
0.1796275013 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 19 |
Hb_098315_090 |
0.1834620612 |
- |
- |
Aldehyde dehydrogenase family 6 member B2 [Morus notabilis] |
| 20 |
Hb_002025_270 |
0.1835833142 |
- |
- |
PREDICTED: uncharacterized protein At4g15970 isoform X1 [Jatropha curcas] |