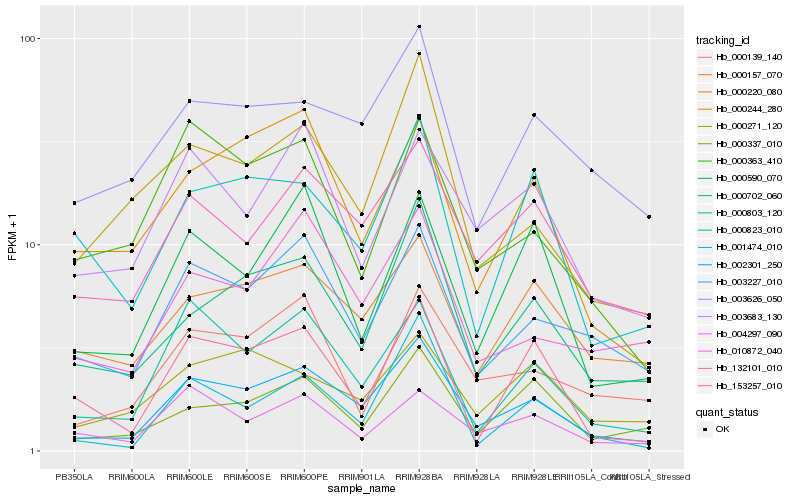
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002301_250 |
0.0 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a [Jatropha curcas] |
| 2 |
Hb_132101_010 |
0.128661714 |
- |
- |
hypothetical protein B456_007G078100 [Gossypium raimondii] |
| 3 |
Hb_000702_060 |
0.13144078 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000803_120 |
0.1323576059 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_003227_010 |
0.1340394784 |
- |
- |
PREDICTED: dynamin-related protein 1E-like [Jatropha curcas] |
| 6 |
Hb_000244_280 |
0.1442187153 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003626_050 |
0.1514882753 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 8 |
Hb_010872_040 |
0.1517330061 |
- |
- |
hypothetical protein F775_31993 [Aegilops tauschii] |
| 9 |
Hb_000157_070 |
0.152027859 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |
| 10 |
Hb_000220_080 |
0.1556785347 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_003683_130 |
0.1563514172 |
- |
- |
PREDICTED: enolase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000139_140 |
0.1563762672 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 13 |
Hb_000590_070 |
0.156911588 |
- |
- |
PREDICTED: beta-hexosaminidase 1 [Jatropha curcas] |
| 14 |
Hb_000337_010 |
0.1604546893 |
- |
- |
PREDICTED: proteasome activator subunit 4 isoform X2 [Jatropha curcas] |
| 15 |
Hb_153257_010 |
0.1607166008 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 16 |
Hb_000271_120 |
0.163876575 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Nelumbo nucifera] |
| 17 |
Hb_000823_010 |
0.1648603 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 18 |
Hb_000363_410 |
0.1653182436 |
- |
- |
hypothetical protein JCGZ_00955 [Jatropha curcas] |
| 19 |
Hb_004297_090 |
0.1657731506 |
- |
- |
PREDICTED: DNA polymerase epsilon catalytic subunit A [Jatropha curcas] |
| 20 |
Hb_001474_010 |
0.1658976738 |
- |
- |
PREDICTED: synaptotagmin-3 isoform X1 [Jatropha curcas] |