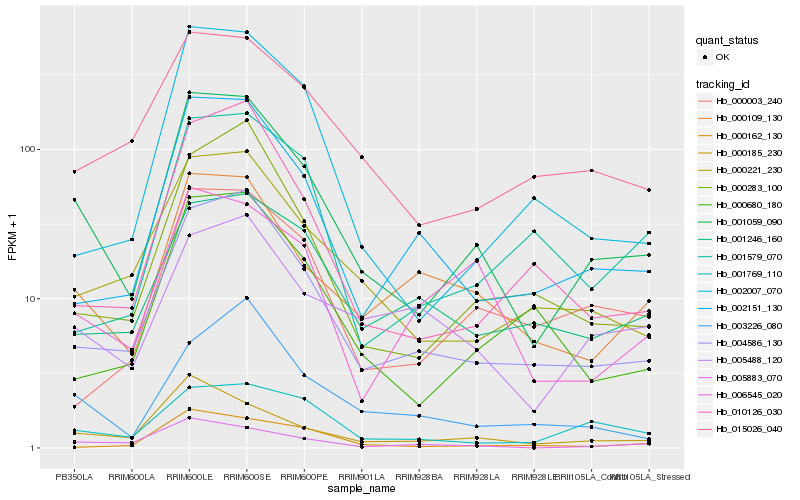
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001059_090 |
0.0 |
- |
- |
PREDICTED: F-box protein At1g61340 [Jatropha curcas] |
| 2 |
Hb_004586_130 |
0.1364795157 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000185_230 |
0.1549307428 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_000221_230 |
0.1580799533 |
transcription factor |
TF Family: Jumonji |
PREDICTED: F-box protein At1g78280 [Jatropha curcas] |
| 5 |
Hb_005883_070 |
0.1678578944 |
- |
- |
PREDICTED: probable myosin-binding protein 5 [Jatropha curcas] |
| 6 |
Hb_002151_130 |
0.1688016755 |
- |
- |
12-oxophytodienoate reductase [Hevea brasiliensis] |
| 7 |
Hb_000109_130 |
0.169148354 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 9 [Jatropha curcas] |
| 8 |
Hb_015026_040 |
0.1739854012 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 9 |
Hb_000283_100 |
0.1771695465 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase DRIP2-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_002007_070 |
0.185587837 |
- |
- |
PREDICTED: serrate RNA effector molecule homolog [Jatropha curcas] |
| 11 |
Hb_010126_030 |
0.1855976396 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000680_180 |
0.1891719768 |
- |
- |
hypothetical protein JCGZ_15248 [Jatropha curcas] |
| 13 |
Hb_001769_110 |
0.190866341 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 14 |
Hb_006545_020 |
0.1926885952 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 15 |
Hb_000003_240 |
0.1933999086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000162_130 |
0.1962846538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003226_080 |
0.1967087826 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5-like [Jatropha curcas] |
| 18 |
Hb_001246_160 |
0.2023002903 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 19 |
Hb_005488_120 |
0.2027561013 |
- |
- |
PREDICTED: DNA polymerase delta subunit 4 [Jatropha curcas] |
| 20 |
Hb_001579_070 |
0.2107219313 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |