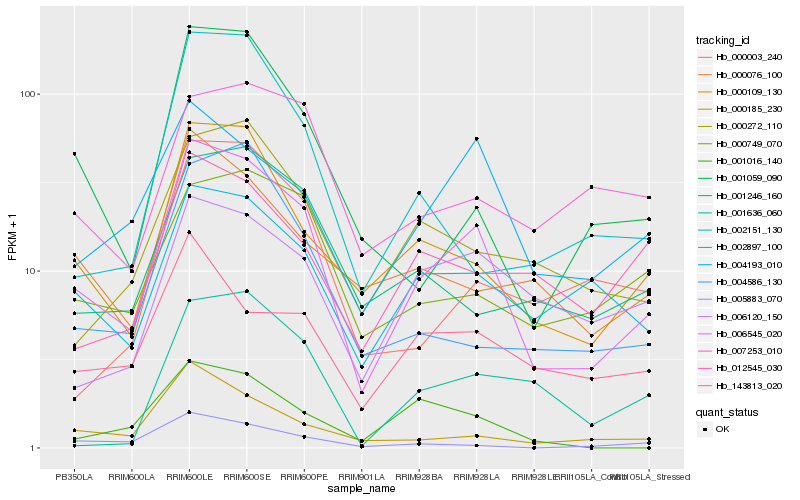
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006545_020 |
0.0 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 2 |
Hb_000109_130 |
0.1588740289 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 9 [Jatropha curcas] |
| 3 |
Hb_002897_100 |
0.1851598546 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 4 |
Hb_000185_230 |
0.1894486567 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 5 |
Hb_004193_010 |
0.1904126942 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |
| 6 |
Hb_001059_090 |
0.1926885952 |
- |
- |
PREDICTED: F-box protein At1g61340 [Jatropha curcas] |
| 7 |
Hb_000272_110 |
0.1955773785 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000749_070 |
0.1974571502 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 9 |
Hb_001246_160 |
0.1985968815 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 10 |
Hb_005883_070 |
0.1986579459 |
- |
- |
PREDICTED: probable myosin-binding protein 5 [Jatropha curcas] |
| 11 |
Hb_000076_100 |
0.2034896713 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1 [Jatropha curcas] |
| 12 |
Hb_143813_020 |
0.2067411419 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 13 |
Hb_004586_130 |
0.2070829976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000003_240 |
0.2093235798 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002151_130 |
0.2102242084 |
- |
- |
12-oxophytodienoate reductase [Hevea brasiliensis] |
| 16 |
Hb_006120_150 |
0.2103966978 |
- |
- |
invertase [Hevea brasiliensis] |
| 17 |
Hb_007253_010 |
0.2168364597 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 18 |
Hb_012545_030 |
0.219067665 |
- |
- |
Polyadenylate-binding protein RBP47C [Glycine soja] |
| 19 |
Hb_001636_060 |
0.2202627502 |
- |
- |
Putative serine/threonine-protein kinase-like protein CCR3 [Glycine soja] |
| 20 |
Hb_001016_140 |
0.2208674975 |
- |
- |
PREDICTED: uncharacterized protein LOC105647117 [Jatropha curcas] |