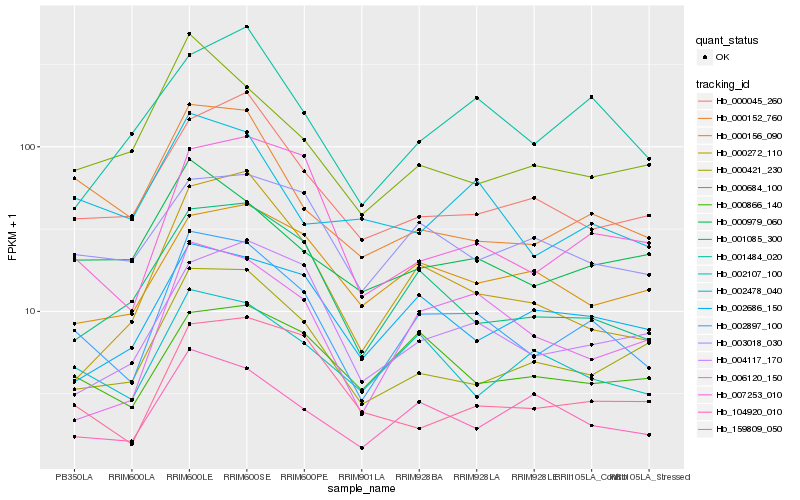
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002897_100 |
0.0 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 2 |
Hb_001085_300 |
0.1384099392 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 3 |
Hb_007253_010 |
0.1440324257 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 4 |
Hb_000152_760 |
0.1506413984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002107_100 |
0.153438495 |
- |
- |
PREDICTED: la-related protein 1C-like isoform X3 [Jatropha curcas] |
| 6 |
Hb_104920_010 |
0.1549059421 |
- |
- |
hypothetical protein VITISV_031859 [Vitis vinifera] |
| 7 |
Hb_000421_230 |
0.1558402487 |
- |
- |
retinal degeneration B beta, putative [Ricinus communis] |
| 8 |
Hb_000045_260 |
0.1563862312 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_006120_150 |
0.1571758165 |
- |
- |
invertase [Hevea brasiliensis] |
| 10 |
Hb_003018_030 |
0.1580056395 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 11 |
Hb_000866_140 |
0.1605429988 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 12 |
Hb_000979_060 |
0.1620356302 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 2-like [Gossypium raimondii] |
| 13 |
Hb_000272_110 |
0.1638995978 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002478_040 |
0.1649616567 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 2C, putative [Ricinus communis] |
| 15 |
Hb_002686_150 |
0.1658241024 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 16 |
Hb_004117_170 |
0.1660248905 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 17 |
Hb_001484_020 |
0.1665071118 |
- |
- |
Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000156_090 |
0.1673461379 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_159809_050 |
0.1676168714 |
- |
- |
AT14A, putative [Ricinus communis] |
| 20 |
Hb_000684_100 |
0.1686912829 |
- |
- |
PREDICTED: cell number regulator 8 [Jatropha curcas] |