| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
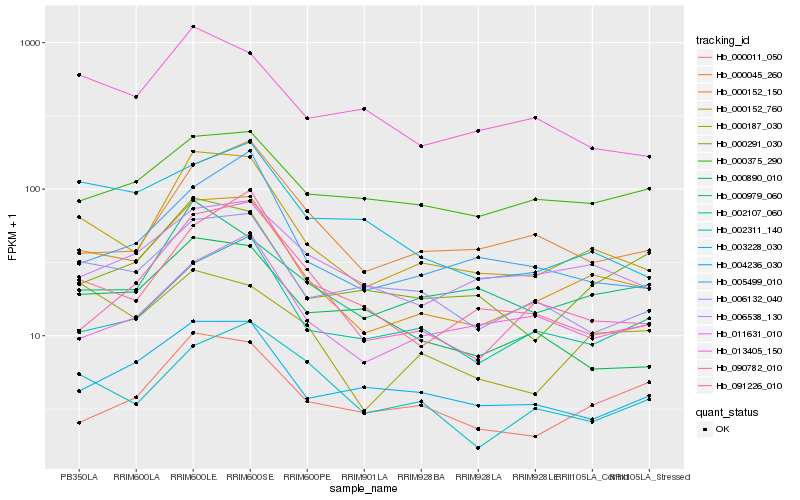
Hb_000152_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648558 [Jatropha curcas] |
| 2 |
Hb_000152_760 |
0.0815837393 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006538_130 |
0.1310127162 |
- |
- |
Uncharacterized protein TCM_009867 [Theobroma cacao] |
| 4 |
Hb_006132_040 |
0.1318317943 |
- |
- |
hypothetical protein JCGZ_14390 [Jatropha curcas] |
| 5 |
Hb_002107_060 |
0.1319625704 |
- |
- |
PREDICTED: uncharacterized protein LOC105635312 [Jatropha curcas] |
| 6 |
Hb_000187_030 |
0.1374593661 |
- |
- |
PREDICTED: cyclin-D4-1-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_003228_030 |
0.142264465 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 8 |
Hb_004236_030 |
0.1436603717 |
- |
- |
unknown [Lotus japonicus] |
| 9 |
Hb_011631_010 |
0.1446828672 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 10 |
Hb_000291_030 |
0.1461051731 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Eucalyptus grandis] |
| 11 |
Hb_090782_010 |
0.1472614184 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 12 |
Hb_005499_010 |
0.1482959143 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 13 |
Hb_000890_010 |
0.148774237 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000375_290 |
0.1488933962 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_013405_150 |
0.1493412222 |
- |
- |
PREDICTED: actin cytoskeleton-regulatory complex protein pan1 [Jatropha curcas] |
| 16 |
Hb_091226_010 |
0.1509495543 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA isoform X1 [Jatropha curcas] |
| 17 |
Hb_002311_140 |
0.1514032612 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 15-like [Jatropha curcas] |
| 18 |
Hb_000045_260 |
0.1523215967 |
- |
- |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_000979_060 |
0.1531888808 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 2-like [Gossypium raimondii] |
| 20 |
Hb_000011_050 |
0.154945033 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |