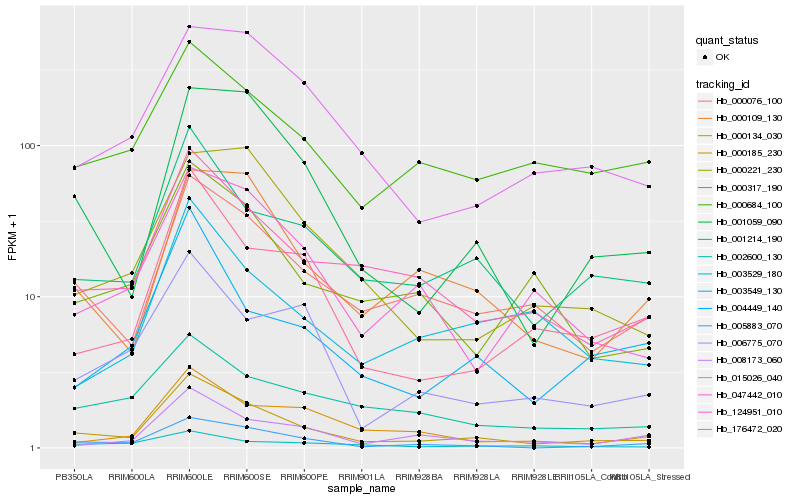
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000185_230 |
0.0 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 2 |
Hb_001214_190 |
0.134885801 |
- |
- |
hypothetical protein JCGZ_25001 [Jatropha curcas] |
| 3 |
Hb_005883_070 |
0.1363547344 |
- |
- |
PREDICTED: probable myosin-binding protein 5 [Jatropha curcas] |
| 4 |
Hb_000076_100 |
0.1420676297 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1 [Jatropha curcas] |
| 5 |
Hb_124951_010 |
0.1434393707 |
- |
- |
chitinase-like protein [Hevea brasiliensis] |
| 6 |
Hb_001059_090 |
0.1549307428 |
- |
- |
PREDICTED: F-box protein At1g61340 [Jatropha curcas] |
| 7 |
Hb_176472_020 |
0.1630307108 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000109_130 |
0.1709284904 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 9 [Jatropha curcas] |
| 9 |
Hb_047442_010 |
0.1722868038 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 10 |
Hb_000317_190 |
0.174403759 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 11 |
Hb_015026_040 |
0.1759624495 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 12 |
Hb_004449_140 |
0.1772481543 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 13 |
Hb_000684_100 |
0.1774624199 |
- |
- |
PREDICTED: cell number regulator 8 [Jatropha curcas] |
| 14 |
Hb_002600_130 |
0.1806273604 |
- |
- |
PREDICTED: uncharacterized protein LOC105646808 [Jatropha curcas] |
| 15 |
Hb_008173_060 |
0.1812109136 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 16 |
Hb_006775_070 |
0.1827651984 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_003529_180 |
0.1843651095 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 18 |
Hb_000221_230 |
0.1849219474 |
transcription factor |
TF Family: Jumonji |
PREDICTED: F-box protein At1g78280 [Jatropha curcas] |
| 19 |
Hb_000134_030 |
0.1859142952 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 3-like [Jatropha curcas] |
| 20 |
Hb_003549_130 |
0.1877054101 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |