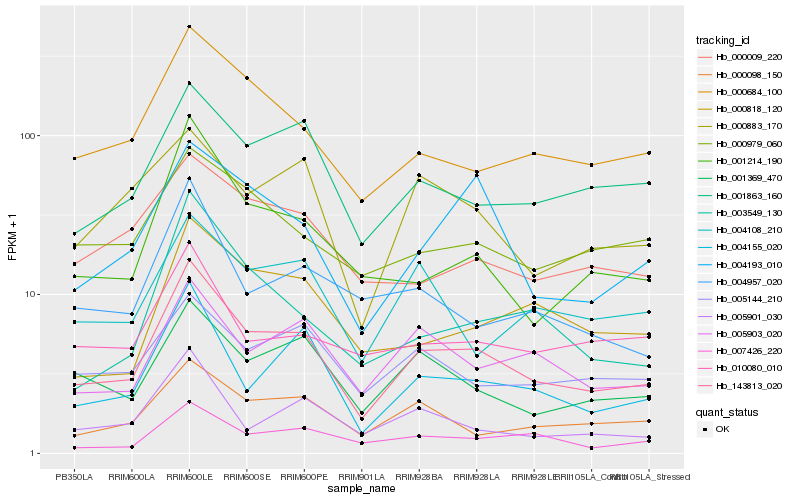
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_143813_020 |
0.0 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 2 |
Hb_004155_020 |
0.1326929396 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 3 |
Hb_000684_100 |
0.1338635375 |
- |
- |
PREDICTED: cell number regulator 8 [Jatropha curcas] |
| 4 |
Hb_001863_160 |
0.1364715519 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 5 |
Hb_001369_470 |
0.1400794395 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKS1 [Jatropha curcas] |
| 6 |
Hb_001214_190 |
0.14290285 |
- |
- |
hypothetical protein JCGZ_25001 [Jatropha curcas] |
| 7 |
Hb_005903_020 |
0.1446399816 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 8 |
Hb_000098_150 |
0.1449128448 |
- |
- |
DNA helicase hus2, putative [Ricinus communis] |
| 9 |
Hb_007426_220 |
0.1450333292 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 10 |
Hb_005901_030 |
0.153539686 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005144_210 |
0.1537623534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000818_120 |
0.1560549326 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004193_010 |
0.1561730497 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |
| 14 |
Hb_003549_130 |
0.1581135423 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 15 |
Hb_004957_020 |
0.1592029605 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000979_060 |
0.159219986 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 2-like [Gossypium raimondii] |
| 17 |
Hb_000009_220 |
0.1598694675 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_004108_210 |
0.1625025481 |
- |
- |
phenazine biosynthesis protein, putative [Ricinus communis] |
| 19 |
Hb_000883_170 |
0.1627866913 |
- |
- |
senescence-associated family protein [Populus trichocarpa] |
| 20 |
Hb_010080_010 |
0.1645544022 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |