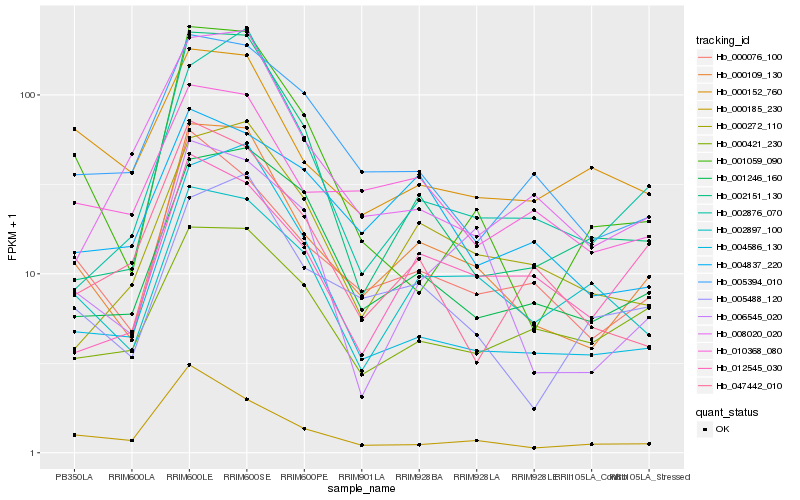
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000109_130 |
0.0 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 9 [Jatropha curcas] |
| 2 |
Hb_000076_100 |
0.1301635806 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1 [Jatropha curcas] |
| 3 |
Hb_005488_120 |
0.1382613266 |
- |
- |
PREDICTED: DNA polymerase delta subunit 4 [Jatropha curcas] |
| 4 |
Hb_001246_160 |
0.1578213405 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 5 |
Hb_006545_020 |
0.1588740289 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 6 |
Hb_004586_130 |
0.1594868404 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_010368_080 |
0.1626259015 |
- |
- |
PREDICTED: uncharacterized protein LOC105630287 [Jatropha curcas] |
| 8 |
Hb_000272_110 |
0.1630272534 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002876_070 |
0.1646583827 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 10 |
Hb_002151_130 |
0.1669132194 |
- |
- |
12-oxophytodienoate reductase [Hevea brasiliensis] |
| 11 |
Hb_005394_010 |
0.1688903655 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 12 |
Hb_001059_090 |
0.169148354 |
- |
- |
PREDICTED: F-box protein At1g61340 [Jatropha curcas] |
| 13 |
Hb_000185_230 |
0.1709284904 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 14 |
Hb_008020_020 |
0.179203217 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000152_760 |
0.1797160449 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002897_100 |
0.1800406408 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 17 |
Hb_047442_010 |
0.1820363226 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 18 |
Hb_012545_030 |
0.1831533307 |
- |
- |
Polyadenylate-binding protein RBP47C [Glycine soja] |
| 19 |
Hb_000421_230 |
0.1833835322 |
- |
- |
retinal degeneration B beta, putative [Ricinus communis] |
| 20 |
Hb_004837_220 |
0.1854218023 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |