| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
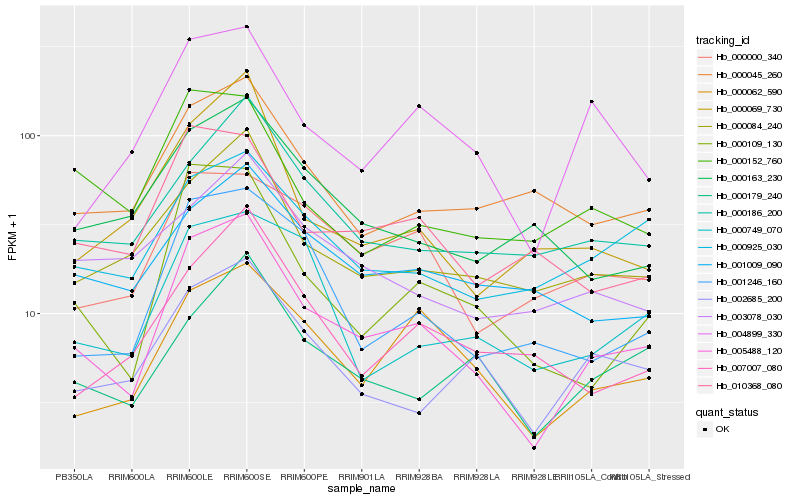
Hb_005488_120 |
0.0 |
- |
- |
PREDICTED: DNA polymerase delta subunit 4 [Jatropha curcas] |
| 2 |
Hb_000109_130 |
0.1382613266 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 9 [Jatropha curcas] |
| 3 |
Hb_000062_590 |
0.1514818891 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_000084_240 |
0.1560393902 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 5 |
Hb_001246_160 |
0.1569072179 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 6 |
Hb_000186_200 |
0.163696561 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 7 |
Hb_002685_200 |
0.1641807788 |
- |
- |
PREDICTED: uncharacterized protein LOC105632583 [Jatropha curcas] |
| 8 |
Hb_004899_330 |
0.1643208529 |
transcription factor |
TF Family: Tify |
plastid jasmonates ZIM-domain protein [Hevea brasiliensis] |
| 9 |
Hb_000179_240 |
0.1663722018 |
- |
- |
PREDICTED: threonine synthase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_010368_080 |
0.1673602483 |
- |
- |
PREDICTED: uncharacterized protein LOC105630287 [Jatropha curcas] |
| 11 |
Hb_000925_030 |
0.1694981585 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 12 |
Hb_000069_730 |
0.1701618704 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000000_340 |
0.1705181687 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor 29-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000045_260 |
0.1759807528 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_001009_090 |
0.1767725914 |
- |
- |
CDK protein [Hevea brasiliensis] |
| 16 |
Hb_000749_070 |
0.1774576956 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 17 |
Hb_000152_760 |
0.1785124873 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007007_080 |
0.1791961589 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000163_230 |
0.1794417158 |
- |
- |
Phytochrome A-associated F-box protein, putative [Ricinus communis] |
| 20 |
Hb_003078_030 |
0.1810622744 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |