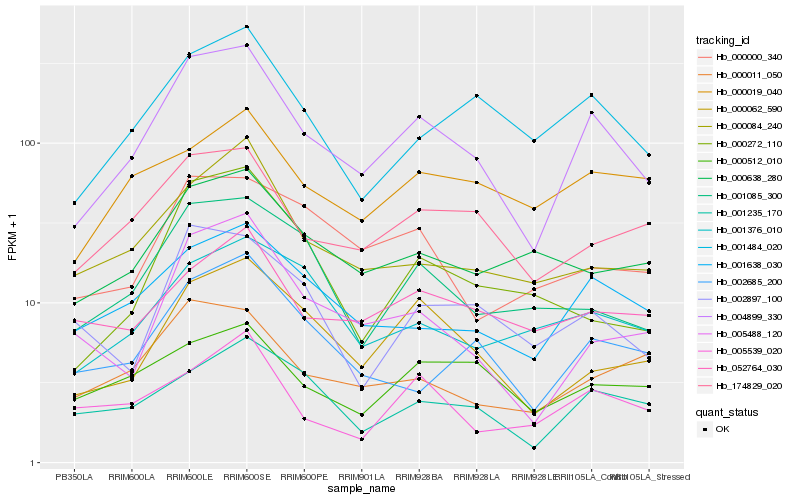
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004899_330 |
0.0 |
transcription factor |
TF Family: Tify |
plastid jasmonates ZIM-domain protein [Hevea brasiliensis] |
| 2 |
Hb_000062_590 |
0.1468977117 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_001638_030 |
0.1497181087 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 4 |
Hb_174829_020 |
0.1513639716 |
transcription factor |
TF Family: C2C2-Dof |
cyclic dof factor 2 [Jatropha curcas] |
| 5 |
Hb_001484_020 |
0.1537544703 |
- |
- |
Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_002685_200 |
0.1626987684 |
- |
- |
PREDICTED: uncharacterized protein LOC105632583 [Jatropha curcas] |
| 7 |
Hb_001235_170 |
0.1628658113 |
- |
- |
PREDICTED: putative glucuronosyltransferase PGSIP8 [Jatropha curcas] |
| 8 |
Hb_001085_300 |
0.163978503 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 9 |
Hb_005488_120 |
0.1643208529 |
- |
- |
PREDICTED: DNA polymerase delta subunit 4 [Jatropha curcas] |
| 10 |
Hb_000084_240 |
0.1661174129 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 11 |
Hb_000011_050 |
0.169739677 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 12 |
Hb_002897_100 |
0.1738000312 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000000_340 |
0.1742874041 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor 29-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000638_280 |
0.1749823834 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 15 |
Hb_001376_010 |
0.1777446539 |
- |
- |
diacylglycerol kinase, alpha, putative [Ricinus communis] |
| 16 |
Hb_000272_110 |
0.1792462103 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000512_010 |
0.1807800329 |
- |
- |
PREDICTED: uncharacterized protein LOC105650728 [Jatropha curcas] |
| 18 |
Hb_000019_040 |
0.183119547 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 3, mitochondrial [Jatropha curcas] |
| 19 |
Hb_005539_020 |
0.183322202 |
- |
- |
- |
| 20 |
Hb_052764_030 |
0.1841440093 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |