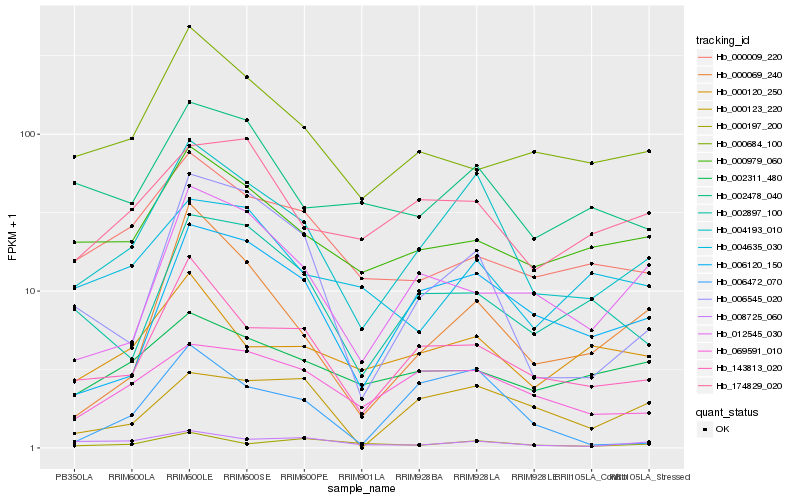
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004193_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |
| 2 |
Hb_143813_020 |
0.1561730497 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 3 |
Hb_006120_150 |
0.1680504756 |
- |
- |
invertase [Hevea brasiliensis] |
| 4 |
Hb_069591_010 |
0.1861246238 |
- |
- |
- |
| 5 |
Hb_002478_040 |
0.188486033 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 2C, putative [Ricinus communis] |
| 6 |
Hb_006545_020 |
0.1904126942 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 7 |
Hb_000979_060 |
0.1922101016 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 2-like [Gossypium raimondii] |
| 8 |
Hb_000120_250 |
0.20001336 |
- |
- |
PREDICTED: uncharacterized protein LOC105630152 isoform X1 [Jatropha curcas] |
| 9 |
Hb_008725_060 |
0.2012157839 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 10 |
Hb_002897_100 |
0.2013223512 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 11 |
Hb_174829_020 |
0.2014403449 |
transcription factor |
TF Family: C2C2-Dof |
cyclic dof factor 2 [Jatropha curcas] |
| 12 |
Hb_000009_220 |
0.2018635057 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_000069_240 |
0.2045514764 |
- |
- |
PREDICTED: protein DEFECTIVE IN MERISTEM SILENCING 3-like [Jatropha curcas] |
| 14 |
Hb_012545_030 |
0.2081970408 |
- |
- |
Polyadenylate-binding protein RBP47C [Glycine soja] |
| 15 |
Hb_004635_030 |
0.2089433887 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 16 |
Hb_006472_070 |
0.2091906084 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 17 |
Hb_000684_100 |
0.2096148538 |
- |
- |
PREDICTED: cell number regulator 8 [Jatropha curcas] |
| 18 |
Hb_000197_200 |
0.2119691626 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 19 |
Hb_002311_480 |
0.2126215615 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000123_220 |
0.2142975529 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_18434 [Jatropha curcas] |