| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
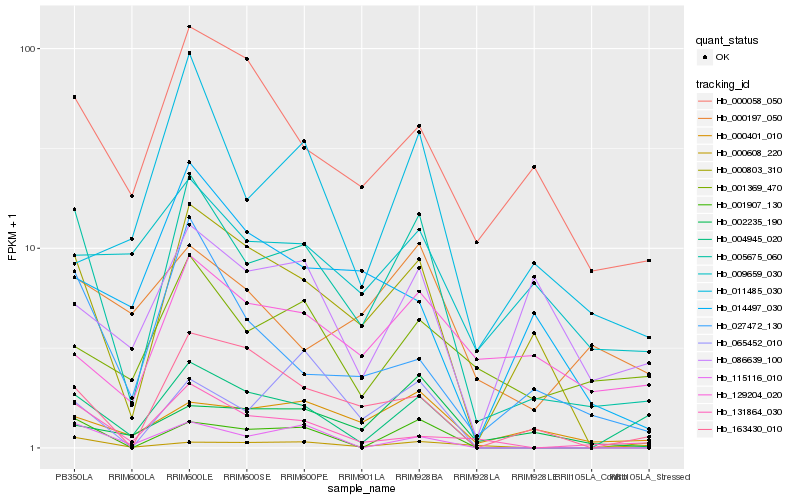
Hb_005675_060 |
0.0 |
- |
- |
PREDICTED: protein NIM1-INTERACTING 2 [Jatropha curcas] |
| 2 |
Hb_000803_310 |
0.1762883204 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105648332 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004945_020 |
0.2362552562 |
- |
- |
PREDICTED: probable calcium-binding protein CML30 [Vitis vinifera] |
| 4 |
Hb_001907_130 |
0.2366180921 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 5 |
Hb_027472_130 |
0.2403693737 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 6 |
Hb_000401_010 |
0.2424592657 |
- |
- |
SAB, putative [Ricinus communis] |
| 7 |
Hb_131864_030 |
0.242906293 |
transcription factor |
TF Family: C2H2 |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_163430_010 |
0.2446312814 |
- |
- |
Polyneuridine-aldehyde esterase precursor, putative [Ricinus communis] |
| 9 |
Hb_000058_050 |
0.2506264474 |
- |
- |
PREDICTED: uncharacterized protein LOC105640877 [Jatropha curcas] |
| 10 |
Hb_115116_010 |
0.2526283729 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 11 |
Hb_001369_470 |
0.2701652509 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKS1 [Jatropha curcas] |
| 12 |
Hb_000608_220 |
0.2722134129 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 13 |
Hb_011485_030 |
0.2727976594 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |
| 14 |
Hb_065452_010 |
0.2750504957 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_129204_020 |
0.276769765 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 4 [Jatropha curcas] |
| 16 |
Hb_002235_190 |
0.278535978 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000197_050 |
0.279398816 |
- |
- |
AGPase large subunit protein [Hevea brasiliensis] |
| 18 |
Hb_009659_030 |
0.2797247715 |
- |
- |
PREDICTED: beta-galactosidase 17 [Jatropha curcas] |
| 19 |
Hb_014497_030 |
0.2887045124 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein BUBR1 [Jatropha curcas] |
| 20 |
Hb_086639_100 |
0.2889993449 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105646787 [Jatropha curcas] |