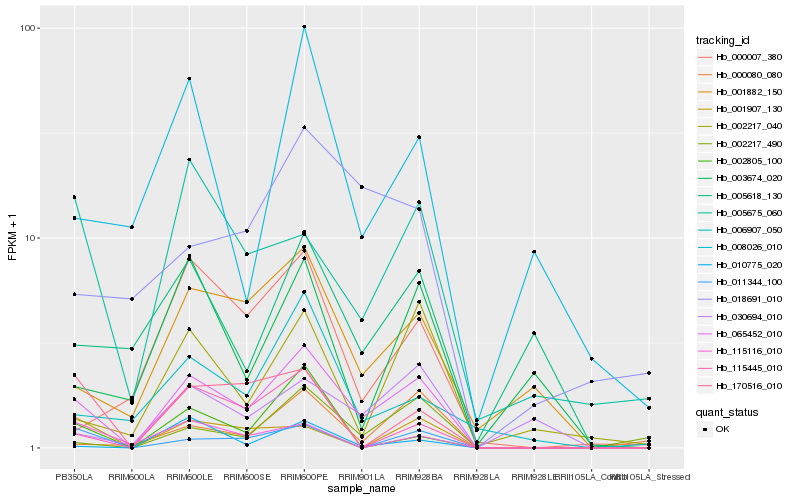
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_065452_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001882_150 |
0.2436051262 |
transcription factor |
TF Family: mTERF |
- |
| 3 |
Hb_011344_100 |
0.2555554043 |
- |
- |
- |
| 4 |
Hb_115445_010 |
0.2692430328 |
- |
- |
- |
| 5 |
Hb_002217_490 |
0.2722498898 |
- |
- |
- |
| 6 |
Hb_030694_010 |
0.2748392318 |
- |
- |
PREDICTED: endoglucanase 25-like [Solanum tuberosum] |
| 7 |
Hb_005675_060 |
0.2750504957 |
- |
- |
PREDICTED: protein NIM1-INTERACTING 2 [Jatropha curcas] |
| 8 |
Hb_002217_040 |
0.2752245102 |
- |
- |
PREDICTED: boron transporter 1-like [Jatropha curcas] |
| 9 |
Hb_170516_010 |
0.2801443748 |
- |
- |
hypothetical protein MTR_001s0023 [Medicago truncatula] |
| 10 |
Hb_006907_050 |
0.2807877538 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_001907_130 |
0.2820368668 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000007_380 |
0.2837258101 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |
| 13 |
Hb_002805_100 |
0.2883227263 |
- |
- |
Receptor-like protein 12 [Morus notabilis] |
| 14 |
Hb_005618_130 |
0.2883335724 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 15 |
Hb_008026_010 |
0.2908513584 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 9 [Jatropha curcas] |
| 16 |
Hb_115116_010 |
0.2919104197 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 17 |
Hb_003674_020 |
0.2939547404 |
- |
- |
PREDICTED: DNA replication licensing factor MCM6 [Jatropha curcas] |
| 18 |
Hb_010775_020 |
0.2953001804 |
transcription factor |
TF Family: ERF |
Floral homeotic protein APETALA2, putative [Ricinus communis] |
| 19 |
Hb_000080_080 |
0.2961669534 |
- |
- |
- |
| 20 |
Hb_018691_010 |
0.3002920829 |
- |
- |
- |