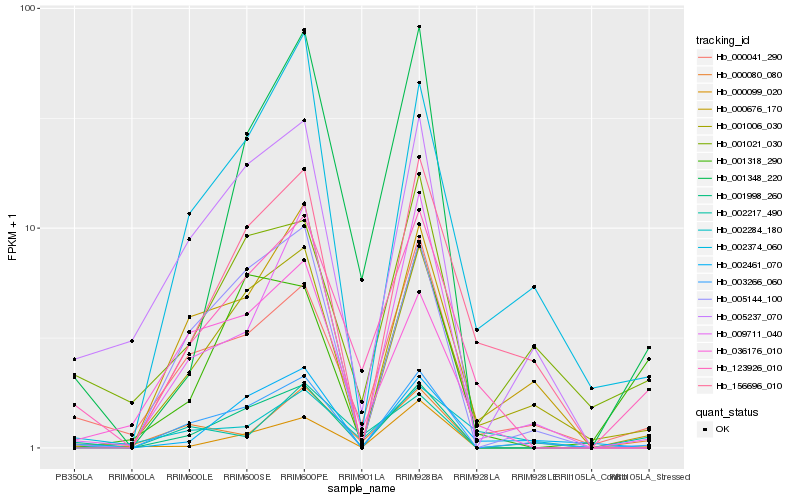
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_123926_010 |
0.0 |
- |
- |
PREDICTED: probable glutathione peroxidase 8-like [Glycine max] |
| 2 |
Hb_000041_290 |
0.1851969377 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 3 |
Hb_001348_220 |
0.216602334 |
- |
- |
PREDICTED: defensin Ec-AMP-D2-like [Nelumbo nucifera] |
| 4 |
Hb_000080_080 |
0.2166126424 |
- |
- |
- |
| 5 |
Hb_001006_030 |
0.2177810917 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002217_490 |
0.2187718917 |
- |
- |
- |
| 7 |
Hb_036176_010 |
0.223098537 |
- |
- |
hypothetical protein CISIN_1g002055mg [Citrus sinensis] |
| 8 |
Hb_001021_030 |
0.2354038918 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 9 |
Hb_002284_180 |
0.2408443577 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 10 |
Hb_002374_060 |
0.2409734504 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 11 |
Hb_003266_060 |
0.241630981 |
- |
- |
PREDICTED: basic 7S globulin-like [Populus euphratica] |
| 12 |
Hb_000676_170 |
0.2426503333 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 13 |
Hb_156696_010 |
0.242972404 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 14 |
Hb_001998_260 |
0.2462637311 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_009711_040 |
0.2471268205 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 16 |
Hb_001318_290 |
0.2478386373 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 17 |
Hb_005237_070 |
0.2509750589 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 4-like [Populus euphratica] |
| 18 |
Hb_005144_100 |
0.2519527086 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 19 |
Hb_000099_020 |
0.2537860183 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: uncharacterized protein LOC103431057 [Malus domestica] |
| 20 |
Hb_002461_070 |
0.2549924229 |
- |
- |
gibberellin 2-oxidase, putative [Ricinus communis] |