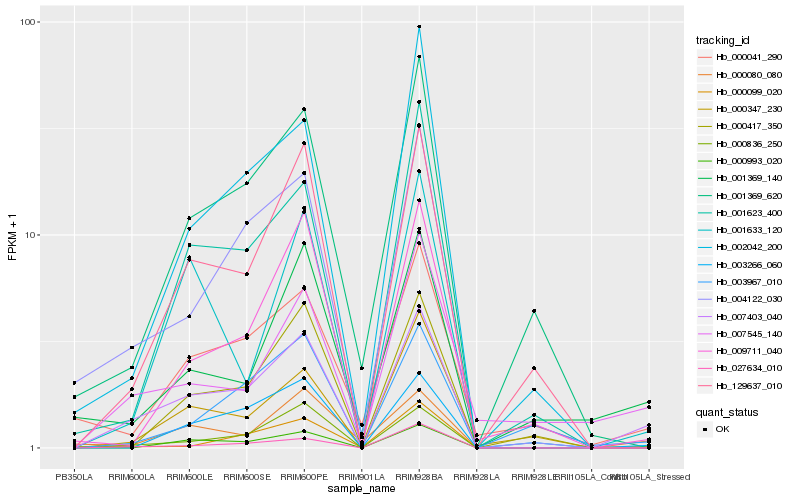
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007403_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000836_250 |
0.1841521079 |
- |
- |
hypothetical protein VITISV_022299 [Vitis vinifera] |
| 3 |
Hb_004122_030 |
0.189754928 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 4-like [Populus euphratica] |
| 4 |
Hb_129637_010 |
0.1902514466 |
- |
- |
PREDICTED: alpha-mannosidase isoform X1 [Populus euphratica] |
| 5 |
Hb_027634_010 |
0.1960519671 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_000080_080 |
0.1990697044 |
- |
- |
- |
| 7 |
Hb_000993_020 |
0.2042090596 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 8 |
Hb_003266_060 |
0.205592014 |
- |
- |
PREDICTED: basic 7S globulin-like [Populus euphratica] |
| 9 |
Hb_001623_400 |
0.206665665 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007545_140 |
0.2092093205 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105141503 [Populus euphratica] |
| 11 |
Hb_000041_290 |
0.2121428996 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 12 |
Hb_000347_230 |
0.2136104814 |
- |
- |
inter-alpha-trypsin inhibitor heavy chain, putative [Ricinus communis] |
| 13 |
Hb_001369_620 |
0.2157179727 |
- |
- |
PREDICTED: protein EXORDIUM-like 3 [Jatropha curcas] |
| 14 |
Hb_000417_350 |
0.2157262707 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_009711_040 |
0.217375168 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 16 |
Hb_001633_120 |
0.2176482371 |
- |
- |
PREDICTED: uncharacterized protein LOC105630199 [Jatropha curcas] |
| 17 |
Hb_002042_200 |
0.2193711491 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_001369_140 |
0.2196379391 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003967_010 |
0.2207199754 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_000099_020 |
0.2227114844 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: uncharacterized protein LOC103431057 [Malus domestica] |