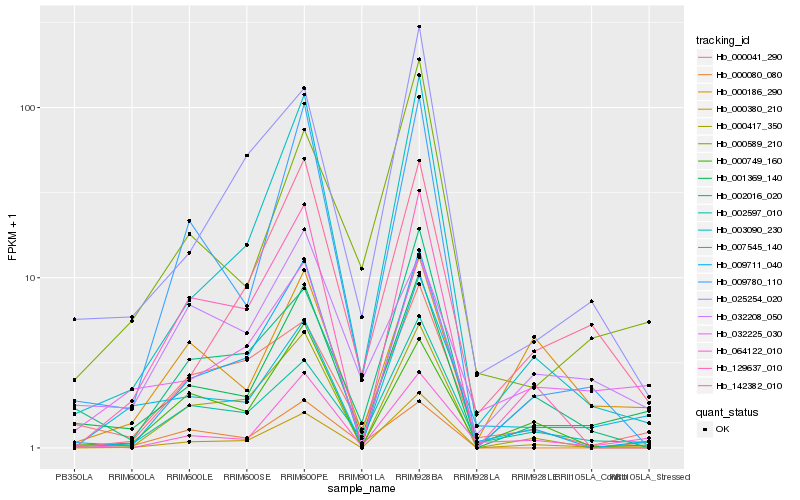
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_064122_010 |
0.151709016 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000380_210 |
0.1717199084 |
- |
- |
Cyclic nucleotide-gated channel 15 [Theobroma cacao] |
| 4 |
Hb_000080_080 |
0.1722710823 |
- |
- |
- |
| 5 |
Hb_032225_030 |
0.1725698785 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 2 [Jatropha curcas] |
| 6 |
Hb_009780_110 |
0.1755664116 |
- |
- |
PREDICTED: protein RSI-1 [Cicer arietinum] |
| 7 |
Hb_002597_010 |
0.1756955485 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Malus domestica] |
| 8 |
Hb_007545_140 |
0.1813692304 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105141503 [Populus euphratica] |
| 9 |
Hb_003090_230 |
0.181967256 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_009711_040 |
0.1846819155 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 11 |
Hb_000041_290 |
0.1917407226 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 12 |
Hb_000749_160 |
0.1972340545 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 13 |
Hb_000589_210 |
0.1986500254 |
- |
- |
PREDICTED: uncharacterized protein LOC105647439 [Jatropha curcas] |
| 14 |
Hb_002016_020 |
0.1990877612 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 15 |
Hb_142382_010 |
0.1994499944 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 16 |
Hb_129637_010 |
0.201835146 |
- |
- |
PREDICTED: alpha-mannosidase isoform X1 [Populus euphratica] |
| 17 |
Hb_000417_350 |
0.2040669194 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000186_290 |
0.2042656301 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 19 |
Hb_032208_050 |
0.2056963466 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 4 isoform X3 [Jatropha curcas] |
| 20 |
Hb_025254_020 |
0.2060414221 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Vitis vinifera] |