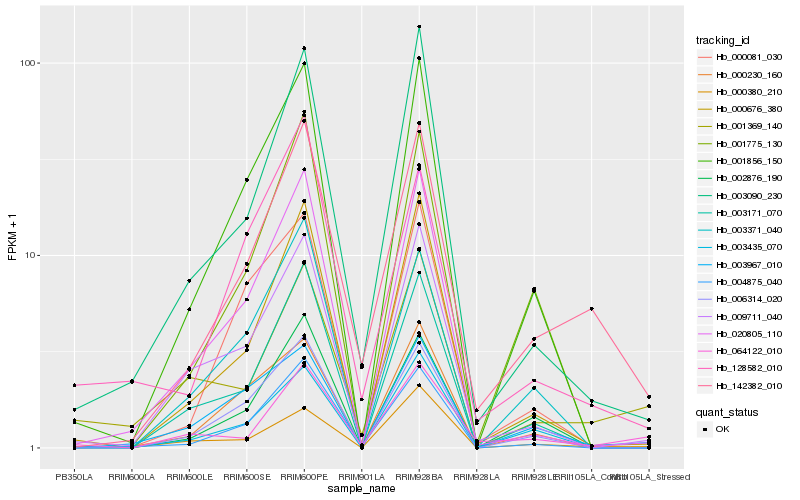
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_142382_010 |
0.0 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 2 |
Hb_003090_230 |
0.1660723352 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000676_380 |
0.175476801 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 4 |
Hb_004875_040 |
0.1774703185 |
- |
- |
PREDICTED: uncharacterized protein LOC105645797 [Jatropha curcas] |
| 5 |
Hb_001856_150 |
0.1873032385 |
- |
- |
- |
| 6 |
Hb_001775_130 |
0.1909409167 |
- |
- |
Prephenate dehydratase [Hevea brasiliensis] |
| 7 |
Hb_020805_110 |
0.1910420683 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 8 |
Hb_064122_010 |
0.1919448245 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X2 [Jatropha curcas] |
| 9 |
Hb_003171_070 |
0.1929640629 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Citrus sinensis] |
| 10 |
Hb_009711_040 |
0.1935103809 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 11 |
Hb_006314_020 |
0.1939826981 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 12 |
Hb_128582_010 |
0.1962618199 |
- |
- |
- |
| 13 |
Hb_001369_140 |
0.1994499944 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000380_210 |
0.1996318927 |
- |
- |
Cyclic nucleotide-gated channel 15 [Theobroma cacao] |
| 15 |
Hb_000081_030 |
0.2004433308 |
- |
- |
BnaC09g05810D [Brassica napus] |
| 16 |
Hb_002876_190 |
0.2014900207 |
transcription factor |
TF Family: GRAS |
hypothetical protein PRUPE_ppa017519mg [Prunus persica] |
| 17 |
Hb_003435_070 |
0.2037811321 |
- |
- |
PREDICTED: probable pectate lyase 12 isoform X3 [Jatropha curcas] |
| 18 |
Hb_003967_010 |
0.2062964785 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_003371_040 |
0.2063030761 |
- |
- |
PREDICTED: uncharacterized protein LOC105650327 [Jatropha curcas] |
| 20 |
Hb_000230_160 |
0.2067324699 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X1 [Jatropha curcas] |