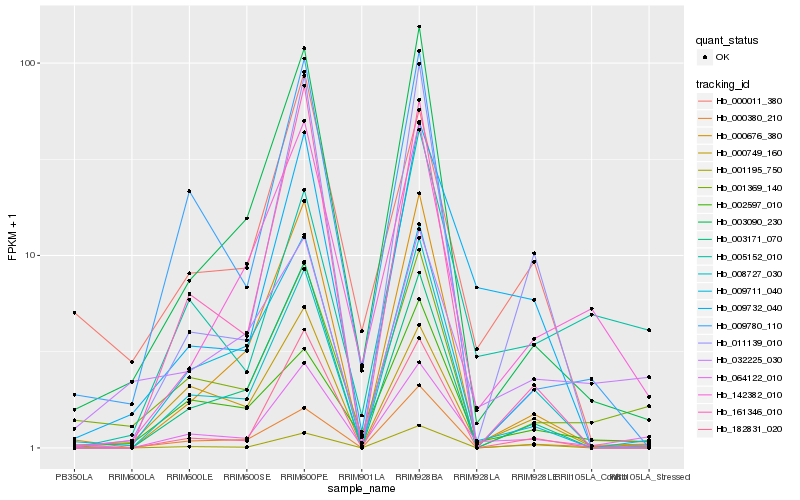
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_064122_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001369_140 |
0.151709016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_009711_040 |
0.1836048301 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 4 |
Hb_000676_380 |
0.1888076599 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 5 |
Hb_003090_230 |
0.1894751444 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_009732_040 |
0.1905912502 |
- |
- |
hypothetical protein POPTR_0006s04880g [Populus trichocarpa] |
| 7 |
Hb_142382_010 |
0.1919448245 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 8 |
Hb_000011_380 |
0.1964152525 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 9 |
Hb_000749_160 |
0.1972669638 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 10 |
Hb_001195_750 |
0.1974502844 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 [Jatropha curcas] |
| 11 |
Hb_000380_210 |
0.1987443359 |
- |
- |
Cyclic nucleotide-gated channel 15 [Theobroma cacao] |
| 12 |
Hb_009780_110 |
0.1998393063 |
- |
- |
PREDICTED: protein RSI-1 [Cicer arietinum] |
| 13 |
Hb_005152_010 |
0.1999418802 |
- |
- |
hypothetical protein JCGZ_22048 [Jatropha curcas] |
| 14 |
Hb_003171_070 |
0.2044764914 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Citrus sinensis] |
| 15 |
Hb_032225_030 |
0.2046551175 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 2 [Jatropha curcas] |
| 16 |
Hb_161346_010 |
0.2100720099 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 20 [Populus euphratica] |
| 17 |
Hb_182831_020 |
0.2105679111 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 18 |
Hb_002597_010 |
0.2119162381 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Malus domestica] |
| 19 |
Hb_008727_030 |
0.2125384845 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_011139_010 |
0.213705354 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |