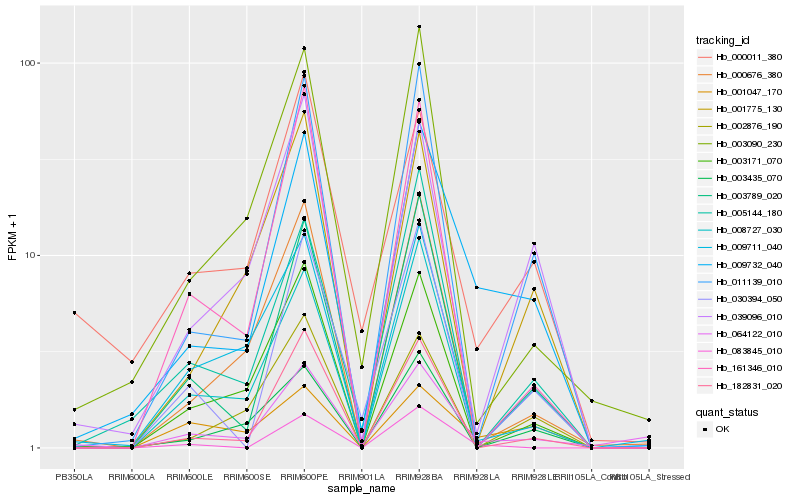
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009732_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s04880g [Populus trichocarpa] |
| 2 |
Hb_011139_010 |
0.1754112994 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_064122_010 |
0.1905912502 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X2 [Jatropha curcas] |
| 4 |
Hb_008727_030 |
0.195305833 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003789_020 |
0.1978812885 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 6 |
Hb_039096_010 |
0.1984477295 |
- |
- |
unknown [Populus trichocarpa] |
| 7 |
Hb_182831_020 |
0.2000476066 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 8 |
Hb_005144_180 |
0.2032497606 |
- |
- |
PREDICTED: protein enabled homolog [Populus euphratica] |
| 9 |
Hb_000676_380 |
0.2035816378 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 10 |
Hb_003435_070 |
0.2041128821 |
- |
- |
PREDICTED: probable pectate lyase 12 isoform X3 [Jatropha curcas] |
| 11 |
Hb_003090_230 |
0.2047162147 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000011_380 |
0.205156903 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 13 |
Hb_083845_010 |
0.2059131037 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 14 |
Hb_001775_130 |
0.2093931926 |
- |
- |
Prephenate dehydratase [Hevea brasiliensis] |
| 15 |
Hb_003171_070 |
0.2142681503 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Citrus sinensis] |
| 16 |
Hb_030394_050 |
0.2168749501 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 17 |
Hb_001047_170 |
0.2193804548 |
- |
- |
PREDICTED: uncharacterized protein LOC105649981 [Jatropha curcas] |
| 18 |
Hb_161346_010 |
0.2198686507 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 20 [Populus euphratica] |
| 19 |
Hb_002876_190 |
0.2218106495 |
transcription factor |
TF Family: GRAS |
hypothetical protein PRUPE_ppa017519mg [Prunus persica] |
| 20 |
Hb_009711_040 |
0.2227314756 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |