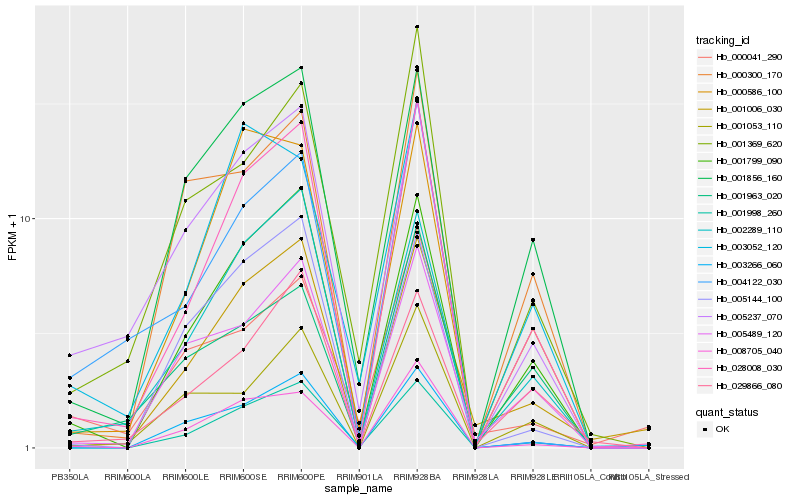
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005237_070 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 4-like [Populus euphratica] |
| 2 |
Hb_002289_110 |
0.1073420725 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 3 |
Hb_001856_160 |
0.1263217074 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001799_090 |
0.1322986829 |
- |
- |
PREDICTED: uclacyanin-2-like [Jatropha curcas] |
| 5 |
Hb_028008_030 |
0.1441517296 |
- |
- |
probable xyloglucan endotransglucosylase/hydrolase protein 23 precursor [Malus domestica] |
| 6 |
Hb_005144_100 |
0.1478627508 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 7 |
Hb_005489_120 |
0.1504978816 |
- |
- |
leucine rich repeat receptor kinase, putative [Ricinus communis] |
| 8 |
Hb_001998_260 |
0.1519755981 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_001369_620 |
0.153780491 |
- |
- |
PREDICTED: protein EXORDIUM-like 3 [Jatropha curcas] |
| 10 |
Hb_004122_030 |
0.1549537867 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 4-like [Populus euphratica] |
| 11 |
Hb_000586_100 |
0.1644725584 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_008705_040 |
0.1675393785 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 13 |
Hb_003266_060 |
0.1682501351 |
- |
- |
PREDICTED: basic 7S globulin-like [Populus euphratica] |
| 14 |
Hb_000300_170 |
0.1695838782 |
- |
- |
PREDICTED: uncharacterized protein LOC105632565 [Jatropha curcas] |
| 15 |
Hb_029866_080 |
0.1704916888 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 16 |
Hb_001053_110 |
0.1707925284 |
- |
- |
PREDICTED: uncharacterized protein LOC105628699 [Jatropha curcas] |
| 17 |
Hb_001006_030 |
0.1722472375 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003052_120 |
0.1756023926 |
- |
- |
hypothetical protein POPTR_0008s10970g [Populus trichocarpa] |
| 19 |
Hb_000041_290 |
0.1759683376 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 20 |
Hb_001963_020 |
0.1769287662 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |