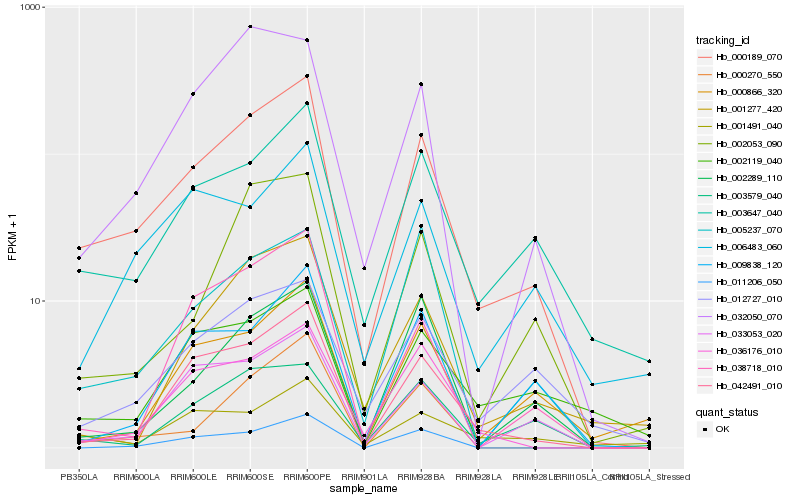
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000189_070 |
0.0 |
transcription factor |
TF Family: AUX/IAA |
Auxin-induced protein 22D, putative [Ricinus communis] |
| 2 |
Hb_042491_010 |
0.1435236784 |
- |
- |
JHL06P13.14 [Jatropha curcas] |
| 3 |
Hb_003647_040 |
0.1457298241 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_033053_020 |
0.1695643381 |
- |
- |
hypothetical protein JCGZ_01204 [Jatropha curcas] |
| 5 |
Hb_012727_010 |
0.1709521374 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 6 |
Hb_036176_010 |
0.1743106553 |
- |
- |
hypothetical protein CISIN_1g002055mg [Citrus sinensis] |
| 7 |
Hb_032050_070 |
0.175444582 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 8 |
Hb_002053_090 |
0.1801784423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_038718_010 |
0.183405987 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 10 |
Hb_005237_070 |
0.186169477 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 4-like [Populus euphratica] |
| 11 |
Hb_002119_040 |
0.1868455699 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 12 |
Hb_003579_040 |
0.1875765226 |
- |
- |
PREDICTED: monoacylglycerol lipase ABHD6 [Jatropha curcas] |
| 13 |
Hb_011206_050 |
0.1879614498 |
- |
- |
hypothetical protein POPTR_0011s02840g [Populus trichocarpa] |
| 14 |
Hb_006483_060 |
0.1885665885 |
- |
- |
PREDICTED: protein RALF-like 34 [Jatropha curcas] |
| 15 |
Hb_001277_420 |
0.1886015253 |
- |
- |
PREDICTED: protein lin-12-like isoform X2 [Populus euphratica] |
| 16 |
Hb_002289_110 |
0.1917137362 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 17 |
Hb_000270_550 |
0.192782737 |
- |
- |
- |
| 18 |
Hb_001491_040 |
0.1928454421 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 19 |
Hb_000866_320 |
0.1929179451 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 20 |
Hb_009838_120 |
0.1939323422 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |