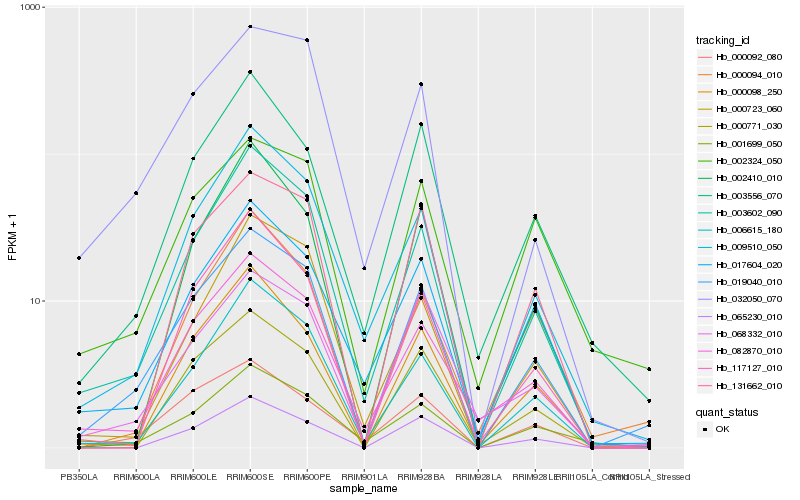
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019040_010 |
0.0 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG7 [Jatropha curcas] |
| 2 |
Hb_003602_090 |
0.1027296987 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9-like [Jatropha curcas] |
| 3 |
Hb_009510_050 |
0.122054097 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_006615_180 |
0.1231784212 |
- |
- |
hypothetical protein B456_003G103400 [Gossypium raimondii] |
| 5 |
Hb_082870_010 |
0.1239192923 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 6 |
Hb_000771_030 |
0.1319826797 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 7 |
Hb_017604_020 |
0.1342049124 |
- |
- |
hypothetical protein JCGZ_22673 [Jatropha curcas] |
| 8 |
Hb_032050_070 |
0.1354445298 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 9 |
Hb_000098_250 |
0.1371315264 |
- |
- |
UDP-glucose flavonoid 3-O-glucosyltransferase [Medicago truncatula] |
| 10 |
Hb_003556_070 |
0.1372439234 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 11 |
Hb_117127_010 |
0.1374526457 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH106 [Jatropha curcas] |
| 12 |
Hb_000092_080 |
0.1386337845 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_038742 [Vitis vinifera] |
| 13 |
Hb_002324_050 |
0.1392544819 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 14 |
Hb_131662_010 |
0.1413784274 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 15 |
Hb_065230_010 |
0.1429202696 |
- |
- |
PREDICTED: uncharacterized protein LOC100814166 isoform X1 [Glycine max] |
| 16 |
Hb_068332_010 |
0.1456125303 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 17 |
Hb_000723_060 |
0.1488786564 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |
| 18 |
Hb_001699_050 |
0.1488892146 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s11410g [Populus trichocarpa] |
| 19 |
Hb_000094_010 |
0.1506697593 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 20 |
Hb_002410_010 |
0.1514046024 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |