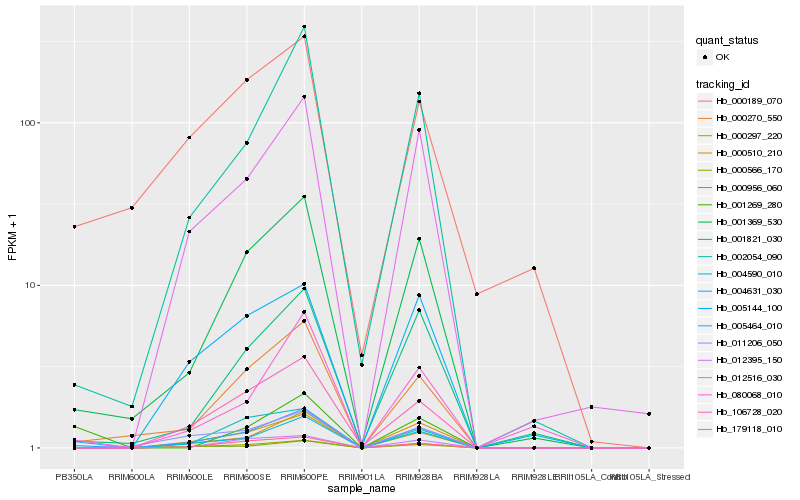
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005464_010 |
0.0 |
- |
- |
Thioredoxin H-type, putative [Ricinus communis] |
| 2 |
Hb_001369_530 |
0.1610041225 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 3 |
Hb_000270_550 |
0.1765704265 |
- |
- |
- |
| 4 |
Hb_004631_030 |
0.1827204446 |
- |
- |
PREDICTED: putative clathrin assembly protein At4g40080 [Jatropha curcas] |
| 5 |
Hb_000510_210 |
0.1832783385 |
- |
- |
PREDICTED: phospholipase A1-Igamma1, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000566_170 |
0.1838906323 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL9 homolog isoform X4 [Jatropha curcas] |
| 7 |
Hb_002054_090 |
0.1898993966 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein-like [Gossypium raimondii] |
| 8 |
Hb_012516_030 |
0.197923552 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 2 {ECO:0000303 |
| 9 |
Hb_106728_020 |
0.1983315071 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_000297_220 |
0.2017648845 |
- |
- |
SAUR family protein [Theobroma cacao] |
| 11 |
Hb_001269_280 |
0.2018867883 |
- |
- |
- |
| 12 |
Hb_001821_030 |
0.2076687054 |
- |
- |
hypothetical protein JCGZ_13357 [Jatropha curcas] |
| 13 |
Hb_000956_060 |
0.2089478794 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_179118_010 |
0.2105961928 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 15 |
Hb_080068_010 |
0.2140154939 |
- |
- |
PREDICTED: basic 7S globulin-like [Jatropha curcas] |
| 16 |
Hb_005144_100 |
0.2227376389 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 17 |
Hb_000189_070 |
0.2236118696 |
transcription factor |
TF Family: AUX/IAA |
Auxin-induced protein 22D, putative [Ricinus communis] |
| 18 |
Hb_011206_050 |
0.224998035 |
- |
- |
hypothetical protein POPTR_0011s02840g [Populus trichocarpa] |
| 19 |
Hb_012395_150 |
0.2260556322 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator-like APRR7 [Jatropha curcas] |
| 20 |
Hb_004590_010 |
0.2288242621 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |