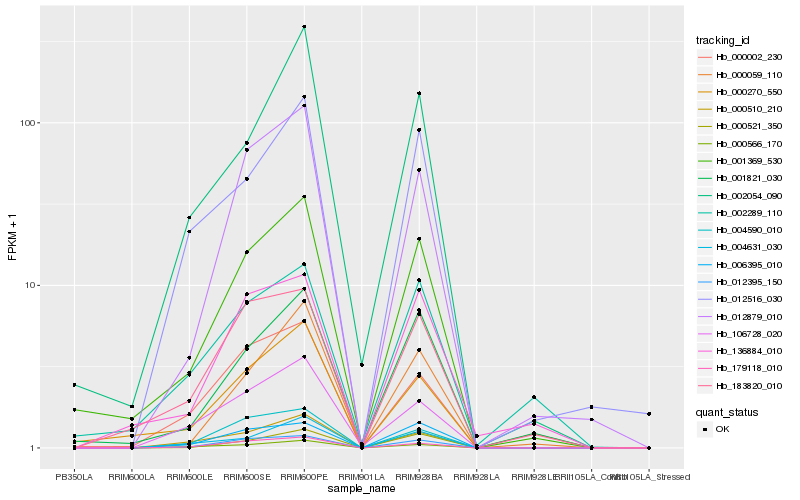
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_530 |
0.0 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 2 |
Hb_000270_550 |
0.0950624834 |
- |
- |
- |
| 3 |
Hb_001821_030 |
0.0979934656 |
- |
- |
hypothetical protein JCGZ_13357 [Jatropha curcas] |
| 4 |
Hb_012395_150 |
0.1337901847 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator-like APRR7 [Jatropha curcas] |
| 5 |
Hb_012879_010 |
0.134836989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_179118_010 |
0.1403030463 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 7 |
Hb_183820_010 |
0.1418608033 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 8 |
Hb_000510_210 |
0.1447088914 |
- |
- |
PREDICTED: phospholipase A1-Igamma1, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_000521_350 |
0.1447728501 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_002054_090 |
0.145528186 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein-like [Gossypium raimondii] |
| 11 |
Hb_000566_170 |
0.1465601205 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL9 homolog isoform X4 [Jatropha curcas] |
| 12 |
Hb_002289_110 |
0.1470287525 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 13 |
Hb_004631_030 |
0.1472273083 |
- |
- |
PREDICTED: putative clathrin assembly protein At4g40080 [Jatropha curcas] |
| 14 |
Hb_136884_010 |
0.1476113287 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 15 |
Hb_000059_110 |
0.1483801273 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus tremula x Populus tremuloides] |
| 16 |
Hb_106728_020 |
0.151970001 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_012516_030 |
0.1529052492 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 2 {ECO:0000303 |
| 18 |
Hb_004590_010 |
0.155521423 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 19 |
Hb_006395_010 |
0.1564565434 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 20 |
Hb_000002_230 |
0.1604425852 |
- |
- |
RING/U-box superfamily protein [Theobroma cacao] |