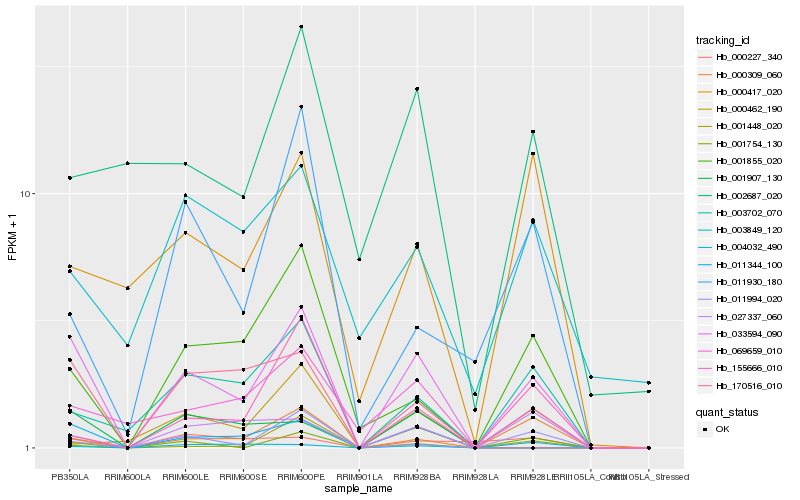
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033594_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_011344_100 |
0.2556180659 |
- |
- |
- |
| 3 |
Hb_003702_070 |
0.2591472474 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_069659_010 |
0.2634093043 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 5 |
Hb_000309_060 |
0.2671202488 |
- |
- |
hypothetical protein JCGZ_09609 [Jatropha curcas] |
| 6 |
Hb_001855_020 |
0.2692783548 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_027337_060 |
0.2785230666 |
- |
- |
- |
| 8 |
Hb_011994_020 |
0.2958544157 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT5-like [Populus euphratica] |
| 9 |
Hb_000417_020 |
0.3015224709 |
- |
- |
PREDICTED: uncharacterized protein LOC105649674 [Jatropha curcas] |
| 10 |
Hb_001448_020 |
0.3016503326 |
- |
- |
hypothetical protein JCGZ_17204 [Jatropha curcas] |
| 11 |
Hb_170516_010 |
0.3066193019 |
- |
- |
hypothetical protein MTR_001s0023 [Medicago truncatula] |
| 12 |
Hb_002687_020 |
0.3071694647 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000227_340 |
0.3078657938 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 14 |
Hb_155666_010 |
0.3094830977 |
- |
- |
hypothetical protein JCGZ_01214 [Jatropha curcas] |
| 15 |
Hb_001754_130 |
0.3096604187 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 16 |
Hb_001907_130 |
0.3135418135 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000462_190 |
0.3158321721 |
- |
- |
PREDICTED: thaumatin-like protein 1 [Jatropha curcas] |
| 18 |
Hb_011930_180 |
0.3159021127 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003849_120 |
0.3172871428 |
- |
- |
PREDICTED: uncharacterized protein LOC100264440 [Vitis vinifera] |
| 20 |
Hb_004032_490 |
0.3174304364 |
- |
- |
PREDICTED: uncharacterized protein LOC100853401 [Vitis vinifera] |