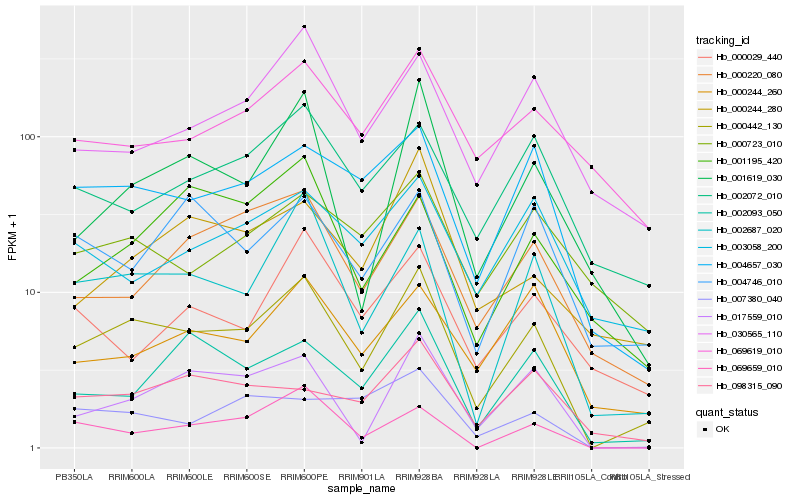
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000442_130 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002687_020 |
0.14771003 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004657_030 |
0.1603890942 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 4 |
Hb_002093_050 |
0.1767276604 |
- |
- |
hypothetical protein POPTR_0008s02940g [Populus trichocarpa] |
| 5 |
Hb_017559_010 |
0.1784723028 |
- |
- |
hypothetical protein JCGZ_06093 [Jatropha curcas] |
| 6 |
Hb_000220_080 |
0.1798148214 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_001619_030 |
0.1801299924 |
- |
- |
annexin, putative [Ricinus communis] |
| 8 |
Hb_002072_010 |
0.1841298991 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_069659_010 |
0.1892859734 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 10 |
Hb_030565_110 |
0.189436266 |
- |
- |
PREDICTED: adenosylhomocysteinase 1 [Jatropha curcas] |
| 11 |
Hb_000244_260 |
0.1895846389 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 12 |
Hb_069619_010 |
0.195674401 |
- |
- |
Aspartic proteinase precursor, putative [Ricinus communis] |
| 13 |
Hb_004746_010 |
0.1961555067 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 14 |
Hb_000244_280 |
0.196795267 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X1 [Jatropha curcas] |
| 15 |
Hb_098315_090 |
0.1975278298 |
- |
- |
Aldehyde dehydrogenase family 6 member B2 [Morus notabilis] |
| 16 |
Hb_003058_200 |
0.198900774 |
- |
- |
PREDICTED: dihydropyrimidinase [Populus euphratica] |
| 17 |
Hb_000723_010 |
0.2009340785 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_007380_040 |
0.2034095079 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 19 |
Hb_001195_420 |
0.2034811007 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X4 [Jatropha curcas] |
| 20 |
Hb_000029_440 |
0.2048192412 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit isoform X2 [Vitis vinifera] |