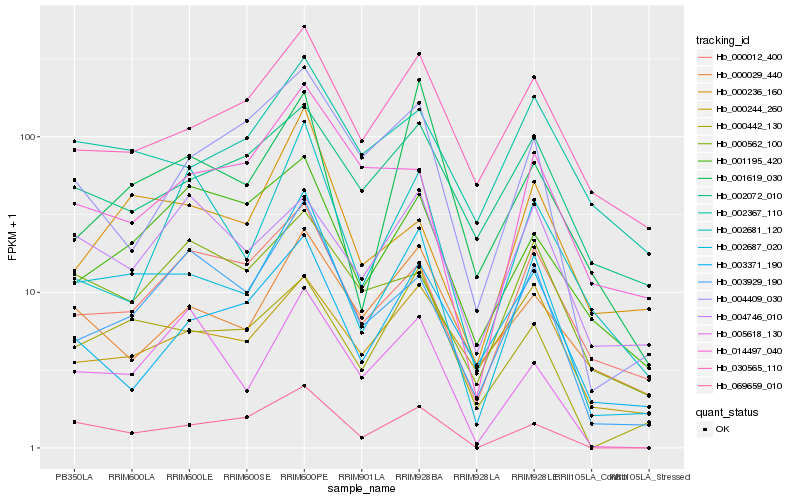
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002687_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_069659_010 |
0.138865841 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 3 |
Hb_000442_130 |
0.14771003 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003929_190 |
0.1652589754 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 5 |
Hb_000012_400 |
0.1724072993 |
- |
- |
PREDICTED: K(+) efflux antiporter 6 [Jatropha curcas] |
| 6 |
Hb_030565_110 |
0.1748264653 |
- |
- |
PREDICTED: adenosylhomocysteinase 1 [Jatropha curcas] |
| 7 |
Hb_002367_110 |
0.1773522098 |
- |
- |
PREDICTED: cinnamyl alcohol dehydrogenase 1 [Sesamum indicum] |
| 8 |
Hb_005618_130 |
0.182479129 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 9 |
Hb_000236_160 |
0.1837294566 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 6 [Jatropha curcas] |
| 10 |
Hb_000029_440 |
0.1843502878 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit isoform X2 [Vitis vinifera] |
| 11 |
Hb_002072_010 |
0.1889980801 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001195_420 |
0.19151872 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X4 [Jatropha curcas] |
| 13 |
Hb_014497_040 |
0.1942604446 |
- |
- |
PREDICTED: calvin cycle protein CP12-3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000562_100 |
0.1942948536 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 15 |
Hb_000244_260 |
0.1959908056 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 16 |
Hb_004409_030 |
0.196691652 |
- |
- |
Glutamine synthetase 1,4 [Theobroma cacao] |
| 17 |
Hb_001619_030 |
0.1980345839 |
- |
- |
annexin, putative [Ricinus communis] |
| 18 |
Hb_003371_190 |
0.1985479346 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 19 |
Hb_004746_010 |
0.2015056543 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 20 |
Hb_002681_120 |
0.2025766422 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g14450 [Jatropha curcas] |