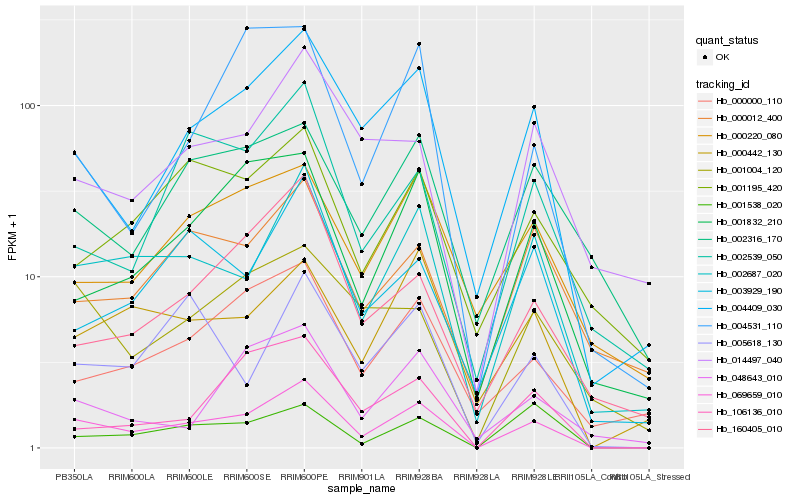
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_069659_010 |
0.0 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 2 |
Hb_002687_020 |
0.138865841 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004409_030 |
0.1535144443 |
- |
- |
Glutamine synthetase 1,4 [Theobroma cacao] |
| 4 |
Hb_000442_130 |
0.1892859734 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001832_210 |
0.1924538181 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 6 |
Hb_000000_110 |
0.1988732895 |
- |
- |
hypothetical protein CISIN_1g0027772mg, partial [Citrus sinensis] |
| 7 |
Hb_001004_120 |
0.2023157272 |
- |
- |
PREDICTED: receptor-like protein kinase HERK 1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001538_020 |
0.2030397099 |
- |
- |
PREDICTED: uncharacterized protein LOC105629608 [Jatropha curcas] |
| 9 |
Hb_106136_010 |
0.203614219 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 10 |
Hb_003929_190 |
0.2050799489 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 11 |
Hb_048643_010 |
0.205732952 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 12 |
Hb_005618_130 |
0.2065796883 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 13 |
Hb_014497_040 |
0.2088923193 |
- |
- |
PREDICTED: calvin cycle protein CP12-3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000012_400 |
0.209782268 |
- |
- |
PREDICTED: K(+) efflux antiporter 6 [Jatropha curcas] |
| 15 |
Hb_002316_170 |
0.2099897984 |
- |
- |
purine permease, putative [Ricinus communis] |
| 16 |
Hb_002539_050 |
0.2104439261 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 17 |
Hb_160405_010 |
0.2107352622 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 18 |
Hb_000220_080 |
0.2107953983 |
- |
- |
kinase, putative [Ricinus communis] |
| 19 |
Hb_004531_110 |
0.2108221572 |
- |
- |
PREDICTED: aquaporin TIP2-1 [Jatropha curcas] |
| 20 |
Hb_001195_420 |
0.2153396804 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X4 [Jatropha curcas] |