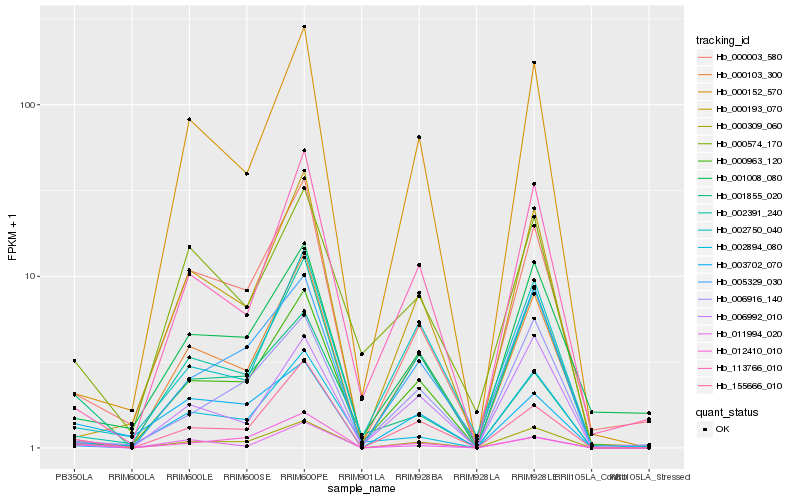
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000309_060 |
0.0 |
- |
- |
hypothetical protein JCGZ_09609 [Jatropha curcas] |
| 2 |
Hb_155666_010 |
0.1675616837 |
- |
- |
hypothetical protein JCGZ_01214 [Jatropha curcas] |
| 3 |
Hb_011994_020 |
0.1780601643 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT5-like [Populus euphratica] |
| 4 |
Hb_002391_240 |
0.1815520558 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_006992_010 |
0.1883207964 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 6 |
Hb_001855_020 |
0.1917705249 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_000003_580 |
0.1922497371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003702_070 |
0.1983268484 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_012410_010 |
0.1987022731 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 10 |
Hb_113766_010 |
0.2071563154 |
- |
- |
plasma membrane H+ ATPase, partial [Nicotiana plumbaginifolia] |
| 11 |
Hb_000103_300 |
0.2074603981 |
- |
- |
PREDICTED: perakine reductase-like [Jatropha curcas] |
| 12 |
Hb_006916_140 |
0.2089956598 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 13 |
Hb_000193_070 |
0.2104578039 |
- |
- |
hypothetical protein JCGZ_25373 [Jatropha curcas] |
| 14 |
Hb_002894_080 |
0.2105946546 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000574_170 |
0.2118448042 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump-like [Jatropha curcas] |
| 16 |
Hb_002750_040 |
0.211926694 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 17 |
Hb_001008_080 |
0.212164053 |
- |
- |
PREDICTED: acyl-protein thioesterase 2 [Jatropha curcas] |
| 18 |
Hb_000963_120 |
0.2141406055 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000152_570 |
0.2143813603 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 20 |
Hb_005329_030 |
0.2154087691 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |