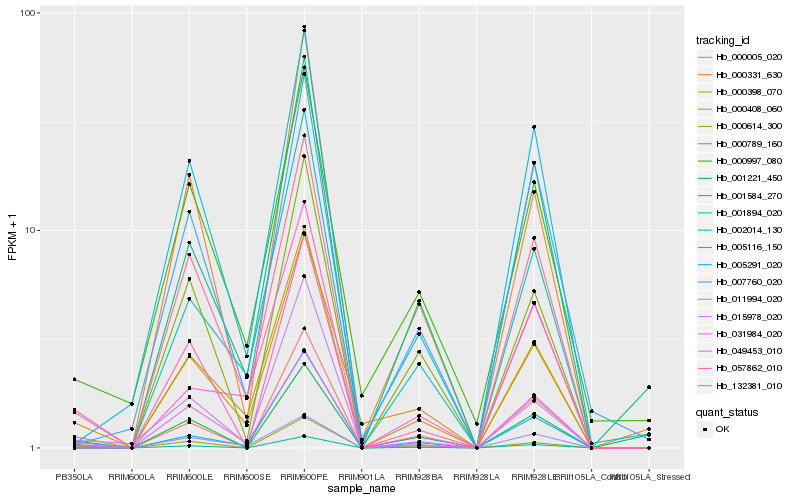
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001894_020 |
0.0 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP630 [Jatropha curcas] |
| 2 |
Hb_000005_020 |
0.1264284962 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_011994_020 |
0.155599121 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT5-like [Populus euphratica] |
| 4 |
Hb_000408_060 |
0.1604530628 |
- |
- |
hypothetical protein POPTR_0018s11360g [Populus trichocarpa] |
| 5 |
Hb_132381_010 |
0.1671720808 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 6 |
Hb_015978_020 |
0.175561826 |
- |
- |
Multidrug resistance protein ABC transporter family protein, putative [Theobroma cacao] |
| 7 |
Hb_000789_160 |
0.1760317718 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: leucine-rich repeat extensin-like protein 4 [Jatropha curcas] |
| 8 |
Hb_000331_630 |
0.1761051294 |
- |
- |
hypothetical protein B456_008G203900 [Gossypium raimondii] |
| 9 |
Hb_049453_010 |
0.1774786691 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_000398_070 |
0.1794404119 |
- |
- |
hypothetical protein VITISV_011880 [Vitis vinifera] |
| 11 |
Hb_000614_300 |
0.180864995 |
- |
- |
PREDICTED: BAHD acyltransferase At5g47980-like [Jatropha curcas] |
| 12 |
Hb_007760_020 |
0.1815632696 |
- |
- |
PREDICTED: expansin-A1 [Populus euphratica] |
| 13 |
Hb_001221_450 |
0.1845667356 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_057862_010 |
0.1850198879 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_031984_020 |
0.1853668913 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001584_270 |
0.1865542291 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000997_080 |
0.1881670162 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005116_150 |
0.1890200452 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002014_130 |
0.1902213836 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005291_020 |
0.1902595428 |
- |
- |
conserved hypothetical protein [Ricinus communis] |